Microsatellite Instability and Tumor Mutational Burden Testing - CAM 342
Description
Tumor mutational burden (TMB) is the genetic characteristic of non-inherited mutations within tumor tissue, often reported as the total number of DNA mutations per one million bases (megabase). Original studies calculated TMB based on whole-exome sequencing and reported TMB as the number of mutations that exist within the exome. However, TMB testing has expanded to targeted gene sequencing panels that do not cover the entire exome. TMB may serve as a biomarker to identify patients likely to have a favorable response to immunotherapy, as high TMB levels correlate with objective response rates to immunotherapy in several different cancer types (Ritterhouse, 2019; C. Willis et al., 2019).
Microsatellites are short, repetitive segments of DNA that are highly prone to mutation. Microsatellite instability (MSI) in tumor DNA is defined as the presence of alternate sized repetitive DNA sequences that are not present in the corresponding germline DNA (Nojadeh et al., 2018). Tumors with high microsatellite instability (MSI-H) are more immunogenic and may therefore respond to drugs that activate the immune system.
Regulatory Status
Many labs have developed specific tests that they must validate and perform in house. These laboratory-developed tests (LDTs) are regulated by the Centers for Medicare & Medicaid Services (CMS) as high-complexity tests under the Clinical Laboratory Improvement Amendments of 1988 (CLIA ’88). LDTs are not approved or cleared by the U.S. Food and Drug Administration; however, FDA clearance or approval is not currently required for clinical use.
On Nov. 30, 2017, the FDA approved FoundationOne CDx™ (Foundation Medicine, Inc.). This device is a next generation sequencing based in vitro diagnostic device for detection of substitutions, insertion, and deletion alterations (indels), and copy number alterations (CNAs) in 324 genes and select gene rearrangements, as well as genomic signatures including microsatellite instability (MSI) and tumor mutational burden (TMB) using DNA isolated from formalin-fixed paraffin embedded (FFPE) tumor tissue specimens. The test is intended as a companion diagnostic to identify patients who may benefit from treatment with the targeted therapies in accordance with the approved therapeutic product labeling. Additionally, F1CDx is intended to provide tumor mutation profiling to be used by qualified health care professionals in accordance with professional guidelines in oncology for patients with solid malignant neoplasms. The F1CDx assay is a single-site assay performed at Foundation Medicine, Inc. (FDA, 2017c).
On Aug. 26, 2020, the FDA approved FoundationOne® Liquid CDx (Foundation Medicine, Inc.). This device is a qualitative next generation sequencing based in vitro diagnostic test that uses targeted high throughput hybridization-based capture technology to detect and report substitutions, insertions, and deletions (indels) in 311 genes, including rearrangements and copy number losses only in BRCA1 and BRCA2, utilizing circulating cell-free DNA (cfDNA) isolated from plasma derived from anti-coagulated peripheral whole blood of cancer patients. This device is intended to provide tumor mutation profiling for substitutions and indels to be used by qualified health care professionals in accordance with professional guidelines in oncology for patients with solid malignant neoplasms (FDA, 2020b).
Policy
- For individuals being considered for immune checkpoint inhibitor (ICI) targeted therapy, tissue-based tumor mutational burden (TMB) and/or microsatellite instability (MSI) testing (Note 1) for the solid tumors outlined in Note 2 is considered MEDICALLY NECESSARY.
- When tissue-based testing is infeasible (i.e., quantity not sufficient for tissue-based test or invasive biopsy is medically contraindicated) for an individual being considered for ICI targeted therapy, liquid biopsy based TMB and/or MSI testing for the solid tumors outlined in Note 2 is considered MEDICALLY NECESSARY.
The following does not meet coverage criteria due to a lack of available published scientific literature confirming that the test(s) is/are required and beneficial for the diagnosis and treatment of an individual’s illness:
- TMB and/or MSI testing for all other tumors and in any other situations not listed above is considered NOT MEDICALLY NECESSARY.
NOTES:
Note 1: NGS panels designed to provide a TMB score are allowed with no restrictions on the number of genes being tested, so long as the panel is designed for TMB assessment.
Note 2: Table of all solid tumors that will be allowed for TMB or MSI testing.
| Condition |
TMB/MSI Targeted Therapy Indications |
| Ampullary adenocarcinoma |
MSI/MMR and TMB testing are recommended for individuals with locally advanced/metastatic adenocarcinoma who are candidates for anti-cancer therapy. |
| Biliary tract cancers (gallbladder, intrahepatic cholangiocarcinoma [CCA], extrahepatic CCA) |
MSI/MMR and TMB testing are recommended for unresectable or metastatic biliary tract cancers (gallbladder, intrahepatic cholangiocarcinoma [CCA], extrahepatic CCA). |
| Bone cancers (metastatic chondrosarcoma, chordoma, Ewing sarcoma, and osteosarcoma, NOT Giant Cell Tumor of Bone) |
Recommended to consider TMB or MSI testing to inform treatment. |
| Breast cancer |
TMB and MSI/dMMR are recommended to guide treatment options for individuals with recurrent unresectable (local or regional) or stage IV (M1) disease. |
| Cervical cancers (squamous cell carcinoma, adenocarcinoma, or adenosquamous carcinoma) |
MSI testing is recommended for recurrent, progressive, or metastatic squamous cell carcinoma, adenocarcinoma, or adenosquamous carcinoma. Comprehensive molecular profiling should be considered for squamous cell carcinoma, adenocarcinoma, or adenosquamous carcinoma. |
| Colon cancer |
MSI/MMR testing advised in all newly diagnosed patients, as a workup in pedunculated or sessile colon polylps (adenoma), in colon cancer appropriate for resection (non-metastatic), in documented metachronous metastases by CT, MRI, and/or biopsy, and in suspected or proven metastatic synchronous adenocarcinoma (any T, any N, M1) if not already done. Stage II MSI-H patients may have a good prognosis and do not benefit from 5-FU adjuvant therapy. Adjuvant therapy should not be given to patients with low-risk stage II MSI-H tumors. MMR/MSI testing should be done during initial workup to help with the diagnosis of Lynch syndrome and inform treatment decision-making if adjuvant therapy is later indicated. |
| Esophageal and Esophagogastric junction (EGJ) cancers |
MSI/MMR is recommended as a universal test in all individuals who have been newly diagnosed with esophageal and EGJ cancers. TMB is considered one of the targeted biomarkers for individuals with esophageal or EGJ cancer. |
| Gastric cancers |
TMB and MSI/dMMR testing are recommended for the clinical management of advanced gastric cancer. |
| Head and Neck Cancers (nasopharyngeal, non-nasopharyngeal, salivary gland tumors) |
TMB is recommended to guide treatment options for individuals with recurrent, unresectable, oligometastatic, or metastatic nasopharyngeal cancer. TMB and MSI/dMMR are recommended to guide treatment options for individuals with non-nasopharyngeal cancers (oral cavity [including mucosal lip], oropharynx, hypopharynx, glottic larynx, supraglottic larynx, ethmoid sinus, maxillary sinus, and occult primary). TMB is recommended as part of follow-up assessment for salivary gland tumors for recurrence with distance metastases. TMB and MSI/dMMR are recommended to guide therapy options in recurrent, unresectable, or metastatic salivary gland tumors. |
| Neuroendocrine/Adrenal cancers (Grade 3 neuroendocrine tumors, adrenocortical carcinoma, and for poorly differentiated neuroendocrine carcinoma/large or small cell) |
TMB and MSI testing should be considered for adrenocortical carcinoma and poorly differentiated neuroendocrine carcinoma/large or small cell and TMB testing should be considered for well-differentiated Grade 3 neuroendocrine tumors. TMB and MSI testing are an appropriate evaluation when the tumor type is poorly differentiated neuroendocrine carcinoma, large or small cell carcinoma (other than the lung), or unknown primary (poorly differentiated). |
| Non-small cell lung cancer |
Biomarker panel testing for actionable driver mutations (with or without TMB/MSI) is allowed. |
| Occult primary cancers (cancer of unknown primary [CUP]) |
Suspected metastatic malignancy should undergo TMB determination or MSI/MMR testing during the workup of the tumor. Of note, the population of patients with MSI-H/dMMR occult primary tumors is low. |
| Ovarian cancers (epithelial ovarian, endometrioid carcinoma, low-grade serous carcinoma, fallopian tube, primary peritoneal cancers) |
TMB and MSI/dMMR is recommended as part of validated molecular testing in Stage I, II, III, and IV epithelial ovarian cancer, fallopian tube cancer, and primary peritoneal cancer when disease is recurrent and CA-125 is rising or there is clinical relapse. TMB and MSI/dMMR are recommended for ovarian cancer, fallopian tube cancer, and primary peritoneal cancer as part of tumor molecular analysis in the recurrence setting. MSI/dMMR testing is recommended for all patients with endometrioid carcinoma. TMB and MSI/dMMR are recommended as part of tumor molecular analysis for monitoring and follow up for recurrence in low-grade serous carcinoma. |
| Pancreatic cancer (adenocarcinoma) |
TMB and MSI/dMMR testing is recommended in locally advanced disease when adenocarcinoma is confirmed, in metastatic disease, and in tumors with recurrence after resection. |
| Pediatric central nervous system cancers (high-grade gliomas EXCEPT oligodendroglioma) |
TMB is recommended to guide targeted therapy of recurrent and progressive disease for diffuse high-grade gliomas EXCEPT oligodendroglioma, IDH-mutant and 1p/19q codeleted or astrocytoma IDH-mutant. |
| Penile cancer |
TMB and MSI/dMMR are recommended to guide treatment options for individuals with metastatic or recurrent disease. |
| Prostate cancers (castration-resistant prostate cancer [CRPC], adenocarcinoma) |
TMB and MSI/dMMR testing should be considered when CRPC imaging studies are positive for metastases. Tumor testing recommended in MSI-H or dMMR among patients with metastatic CRPC and may be considered in patients with regional or castration-naïve metastatic prostate cancer. TMB may be considered in patients with metastatic CRPC. |
| Rectal cancers |
Universal MSI/MMR testing is recommended in all newly diagnosed patients with rectal cancer. In addition, MSI/MMR testing is recommended as an initial workup in pedunculated polyps or sessile polyps (adenoma) with invasive cancer, rectal cancer appropriate for resection, and in suspected or proven metastatic synchronous adenocarcinoma (any T, any N, M1) if not previously done. |
| Small bowel cancer (adenocarcinoma) |
TMB testing should be considered as part of the workup for metastatic and/or recurrent adenocarcinoma. Universal MMR or MSI testing is recommended in all newly diagnosed patients with small bowel adenocarcinoma. When not part of the initial workup, MSI testing is recommended as a workup when the tumor is in the duodenum or the jejunum/ileum and should be part of the workup for metastatic and/or recurrent adenocarcinoma. |
| Soft tissue sarcoma |
TMB and MSI/dMMR are recommended to guide treatment options for individuals with unresectable or metastatic soft tissue sarcoma. |
| Testicular cancers (nonseminoma, pure seminoma) |
TMB and MSI testing are recommended as a third-line therapy in nonseminoma and pure seminoma if there is progression after high-dose chemotherapy or other third-line therapies. |
| Thyroid cancer (anaplastic thyroid carcinoma) |
TMB and MSI/dMMR are recommended as part of initial diagnostic molecular testing for anaplastic thyroid carcinoma. |
| Thyroid cancers (papillary carcinoma, follicular carcinoma, and oncocytic carcinoma) |
TMB and MSI/dMMR testing are recommended for papillary carcinoma, follicular carcinoma, and oncocytic carcinoma for advanced, progressive, or threatening disease when tumor is locally recurrent, advanced, and/or metastatic disease that is not amenable to RAI therapy. |
| Thyroid cancer (medullary carcinoma) |
TMB testing is recommended for medullary carcinoma for recurrent or persistent locoregional disease or for symptomatic disease or progression of disease for recurrent or persistent disease with distance metastases. |
| Uterine cancers (endometrial carcinoma, uterine sarcoma) |
MSI testing is recommended if MMR results are equivocal in endometrial carcinoma. TMB testing should be considered for endometrial carcinoma. For sarcoma, comprehensive genomic profiling (at minimum TMB/MSI, and NTRK) in a setting of metastatic disease is informative for predicting rare pan-tumor targeted therapy opportunities. |
| Vulvar cancer (squamous cell carcinoma) |
MSI and TMB testing may be considered for treatment planning purposes in patients with recurrent, progressive, or metastatic disease. |
MSI-H (MSI-high; MSI-positive); NCCN defines TMB-H as a TMB value of ≥10 mutations/megabase
Table of Terminology
| Term |
Definition |
| 5-FU |
5-fluorouracil |
| ALK |
Anaplastic lymphoma kinase |
| ANTI-PD1 |
Anti-programmed cell death 1 |
| BRAF |
B-Raf proto-oncogene, serine/threonine kinase |
| BRCA1 |
Breast cancer 1 |
| BRCA2 |
Breast cancer 2 |
| bTMB |
Blood tumor mutational burden |
| CA-125 |
Cancer antigen 125 |
| CAPEOX |
Capecitabine and oxaliplatin |
| CCA |
Cholangiocarcinoma |
| cfDNA |
Circulating cell-free deoxyribonucleic acid |
| CGP |
Comprehensive genomic profiling |
| CMS |
Centers for Medicare & Medicaid Services |
| CNA |
Copy number alteration |
| CNS |
Central nervous system |
| CNV |
Copy number variation |
| CPS |
Combined positive score |
| CRM |
Circumferential resection margin |
| CRPC |
Castration-resistant prostate cancer |
| CT |
Computerized tomography |
| ctDNA |
Circulating tumor deoxyribonucleic acid |
| CTLA-4 |
Cytotoxic t-lymphocyte-associated protein 4 |
| CUP |
Cancer of unknown primary |
| DCB |
Durable clinical benefit |
| dMMR |
Deoxyribonucleic acid mismatch repair |
| DNA |
Deoxyribonucleic acid |
| DRUP |
Drug rediscovery protocol |
| EGFR |
Epidermal growth factor receptor |
| EGJ |
Esophagogastric junction |
| ESMO |
European Society for Medical Oncology |
| F1CDx |
FoundationOne CDx™ |
| FBXW7 |
F-box and WD40 repeat domain containing-7 |
| FDA |
Food and Drug Administration |
| FFPE |
Formalin-fixed, paraffin embedded |
| FOLFOX |
Leucovorin calcium, fluorouracil, oxaliplatin |
| GEA |
Gastroesophageal adenocarcinoma |
| HLA |
Human leukocyte antigen |
| ICI |
Immune checkpoint inhibitor |
| IHC |
Immunohistochemistry |
| INDELS |
Insertion and deletions |
| iRECIST |
RECIST 1.1 for immune-based therapeutics |
| LCD |
Local coverage determination |
| LDT |
Laboratory-developed test |
| LOH |
Loss of heterozygosity |
| mCRC |
Metastatic colorectal cancer |
| MLH1 |
MutL homolog 1 |
| MMR |
Mismatch repair |
| MRI |
Magnetic resonance imaging |
| MSH2 |
MutS homolog 2 |
| MSH6 |
MutS homolog 6 |
| MSI |
Microsatellite instability |
| MSI-H |
High microsatellite instability |
| MSI-L |
Low microsatellite instability |
| MSK |
Memorial Sloan Kettering Cancer Center |
| MSS |
Microsatellite stable |
| NCCN |
National Comprehensive Cancer Network |
| NCD |
National coverage determination |
| NCI |
National Cancer Institute |
| NDB |
No durable benefit |
| NET |
Neuroendocrine tumor |
| NGS |
Next-generation sequencing |
| NSCLC |
Non-small cell lung carcinoma |
| NTRK |
Neurotrophic tyrosine receptor kinase |
| ORR |
Objective response rate |
| OS |
Overall survival |
| PCR |
Polymerase chain reaction |
| PD-1 |
Programmed cell death protein |
| PD-L1 |
Programmed cell death protein 1 |
| PFS |
Progression-free survival |
| PMS2 |
Mismatch repair endonuclease PMS2 |
| POLD1 |
Deoxyribonucleic acid polymerase delta catalytic subunit |
| POLE1 |
Deoxyribonucleic acid polymerase epsilon catalytic subunit isoform b |
| PS |
Performance status |
| R2 |
Correlation |
| RAI |
Radioactive iodine |
| RAS |
Retrovirus-associated deoxyribonucleic acid sequences |
| RECIST |
Response evaluation criteria in solid tumors |
| RNA |
Ribonucleic acid |
| ROS1 |
Receptor tyrosine kinase |
| SBA |
Small bowel adenocarcinoma |
| SCL |
Small-cell lung cancer |
| SMAD4 |
SMAD family member 4 |
| SNV |
Single nucleotide variant |
| TF |
Tumor fraction |
| TMB |
Tumor mutational burden |
| TMB-H |
Tumor mutational burden – high |
| TML |
Tumor mutational load |
| TRAE |
Treatment-related adverse events |
| TSO500 |
TruSight oncology 500 |
| tTMB |
Tissue tumor mutation burden |
| TTP |
Time to progression |
| WES |
Whole exome sequencing |
| WTS |
Whole transcriptome sequencing |
Rationale
Tumor mutational burden (TMB) is an emerging biomarker associated with predicting the response to immune checkpoint inhibitors (ICIs). ICIs are therapies that have made significant progress in helping to treat certain advanced cancers. ICIs work by releasing the brakes on the immune system’s antitumor response. ICI therapy has proven most effective on tumor types with a high TMB, whereby a high TMB value indicates better treatment outcomes (Yarchoan et al., 2017).
Clinical biomarkers are widely used for making personalized and actionable decisions for cancer treatment. Many mutations result in production of numerous altered peptides—the more mutations present, the larger the number of altered peptides is produced. A subset of these peptides is expressed and processed by the major histocompatibility complex, resulting in neoantigens. These neoantigens can be recognized by the immune system to generate an antitumor response. The recognition of these neoantigens is thought to be largely stochastic and a higher number of DNA mutations means a higher number of candidate peptides being produced, resulting in a greater likelihood of these neoantigens being recognized by the immune system (Ritterhouse, 2019; Yarchoan et al., 2019).
This phenomenon is observed in patients with melanoma, non-small-cell lung carcinoma (NSCLC), small cell lung cancer, bladder cancer, colorectal cancer, microsatellite instability cancers, and pan-tumors who are on inhibitors of programmed cell death protein (PD-1) and the ligand for programmed cell death protein 1 (PD-L1). High TMB has also been associated with improved outcomes in patients on a combination of PD-1/PD-L1 and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) inhibitors (Hellmann, Callahan, et al., 2018; Hellmann, Nathanson, et al., 2018; Le et al., 2015; Merino et al., 2020; Rizvi et al., 2015; Snyder et al., 2014; Van Allen et al., 2015).
Originally, TMB was measured with whole-exome sequencing (WES). In theory, WES is the best measure of TMB, as it covers every coding sequence within the tumor, ensuring that every mutation contributes to the TMB score. Recently, studies have shown that TMB measured using targeted panels which analyze mutations across specific genes (rather than the whole exome) correlates well with TMB measured by WES (Chalmers et al., 2017; Quy et al., 2019; Wu et al., 2019). WES method has limited clinical utility as it is considered costly, has a high turnaround time (a roughly six to eight week sequencing period), and is difficult for many clinical laboratories to use for routine patient care (Ritterhouse, 2019). Hence, targeted next-generation sequencing (NGS)-based panels that focus analysis on a subset of genes implicated in cancer biology offer several advantages for TMB estimation (Garofalo et al., 2016).
There are FDA-approved products for calculating TMB, including the FoundationOne CDx™ (F1CDx) assay and the FoundationOne Liquid CDx assay (Foundation Medicine Inc.) (FDA, 2017c, 2020b), as well as an additional FDA 510(k) authorized assay, MSK-IMPACT (Memorial Sloan Kettering Cancer Center) (FDA, 2017a, 2017b). These tests, referred to collectively as comprehensive genomic profiling (CGP), can identify all types of “molecular alterations (i.e., single nucleotide variants, small and large insertion‐deletion alterations, copy number alterations, and structural variants) in cancer‐related genes, as well as genomic signatures such as microsatellite instability (MSI), loss of heterozygosity [LOH], and TMB” (Klempner et al., 2020). Studies show that TMB calculated from CGP has high concordance with TMB measured from WES. On June 16, 2020, the FDA approved pembrolizumab for the treatment of adult and pediatric patients with a TMB value of greater than 10 mutations per megabase as determined by the FoundationOne CDx™ assay (FDA, 2020a).
Microsatellites are short, repetitive segments of DNA that are highly prone to mutation. The presence of alternate sized repetitive DNA sequences that are not present in the corresponding germline DNA is defined as microsatellite instability (MSI) (Nojadeh et al., 2018). When a tumor has a high degree of microsatellite instability (MSI-H), it is more immunogenic and thus may respond to drugs that activate the immune system, such as ICIs (Zhao et al., 2019).
The National Cancer Institute (NCI) recommends two mononucleotide repeats (BAT-25 and BAT-26) and three dinucleotide repeats (D5S346, D2S123, and D17S250) as the standard sites/markers in panels for MSI testing (Boland et al., 1998). Per the NCI guidelines, when a tumor shows instability in two or more of the five markers, it is defined as MSI-H. However, this definition may vary by source and markers used (NCI, n.d.). When only a single marker shows instability, tumors may be classified as low-frequency MSI (MSI-L) and tumors lacking any instability are often classified as microsatellite stable (MSS) (Boland et al., 1998).
Microsatellite instability has been identified in many cancer types, with the highest prevalence in uterine corpus endometrial carcinoma, colon adenocarcinoma, stomach adenocarcinoma, and rectal adenocarcinoma. In studies by Bonneville et al. (2017) and Zhao et al. (2019), MSI was undetectable (or at the borderline of detection) in 12 out of 39 cancer types (Bonneville et al., 2017; Zhao et al., 2019).
Proprietary Testing
Caris MI Tumor Seek
Caris Life Sciences has developed MI Tumor Seek™, an NGS-based tumor profiling service that combines WES analysis of DNA for mutations, copy number alterations, insertions/deletions, and genomic signatures (MSI, TMB, and LOH) with Whole Transcriptome Sequencing (WTS) analysis for RNA fusions and variant transcripts. WES covers about 23,000 genes with 200-500x depth of coverage, while WTS covers about 23,000 genes with 60 million reads/sample. DNA and RNA are isolated from formalin-fixed paraffin-embedded (FFPE) tumor tissue “with a minimum of 20% malignant origin for DNA and 10% malignant origin for RNA. Needle biopsy is also acceptable (4-6 cores)” (Caris Life Sciences, 2024).
FoundationOne CDx™
FoundationOne CDx™ is a single-site assay that is “a next generation sequencing based in vitro diagnostic device for detection of substitutions, insertion and deletion alterations (indels) and copy number alterations (CNAs) in 324 genes and select gene rearrangements, as well as genomic signatures including microsatellite instability (MSI) and TMB using DNA isolated from formalin-fixed paraffin-embedded (FFPE) tumor tissue specimens.” It is intended for use by health care professionals as a companion diagnostic to identify patients with solid malignant neoplasms who may benefit from treatment with targeted ICIs (FDA, 2017c).
FoundationOne Liquid CDx
The FoundationOne Liquid CDx assay uses circulating cell-free DNA (cfDNA) that is isolated from plasma from patients with solid malignant neoplasms. The assay uses a “single DNA extraction method to obtain cfDNA from plasma from whole blood. Extracted cfDNA undergoes whole-genome shotgun library construction and hybridization-based capture of 324 cancer-related genes. All coding exons of 309 genes are targeted; select intronic or non-coding regions are targeted in BRCA1 and BRCA2 … Hybrid-capture selected libraries are sequenced with deep coverage using the NovaSeq® 6000 platform. Sequence data are processed using a custom analysis pipeline designed to detect genomic alterations, including base substitutions and indels in 311 genes, and copy number variants and genomic rearrangements in BRCA1 and BRCA2. A subset of targeted regions in 75 genes is baited for increased sensitivity” (FDA, 2020b).
Guardant360® TissueNext
Guardant Health offers Guardant360® CDx, a liquid biopsy that provides comprehensive genomic results from a simple blood draw. This test “uses targeted high throughput hybridization-based capture technology for detection of single nucleotide variants (SNVs), insertions and deletions (indels) in 55 genes, copy number amplifications (CNAs) in two (2) genes, and fusions in four (4) genes. Guardant360® CDx utilizes circulating cell-free DNA (cfDNA) from plasma of peripheral whole blood.” It is an FDA-approved liquid biopsy that can be used for tumor mutation profiling across all solid cancers and as a companion diagnostic to identify NSCLC patients who may benefit from Tagrisso® ( ealth inib), RYBREVANT™ (amivantamab-vmjw), or LUMAKRAS™ (sotorasib). To compliment Guardant360 CDx, Guardant Health also offers Guardant360® TissueNext. Guardant360 TissueNext assesses TMB either from an FFPE tumor specimen or from a blood sample initially processed with Guardant360® CDx. If an actionable biomarker is found in a Guardant360® CDx sample, it is then processed with the Guardant360® TissueNext assay (FDA, 2021; Guardant, 2024a, 2024b).
MSK-IMPACT
The MSK-IMPACT assay is a single-site assay performed at Memorial Sloan Kettering Cancer Center (MSKCC) and is designed to compare two samples from a single patient—a normal specimen and a specimen from a solid malignant neoplasm—allowing the assay to detect tumor gene alterations in a broad multi gene panel (FDA, 2017a). MSK-IMPACT analyzes 505 genes chosen for their critical role in the development and behavior of tumors. To “take full advantage of MSK-IMPACT, MSK doctors and researchers developed a knowledge base called OncoKB®. This system includes information about the clinical and biological effects of more than 4,000 genomic changes. That information is based on public databases, scientific literature, and clinical guidelines” (MSKCC, 2024). The assay is intended to provide information on both somatic mutations and microsatellite instability and is not conclusive or prescriptive for labeled use of any specific therapeutic product (FDA, 2017a).
QIAseq TMB
Qiagen offers QIAseq Tumor Mutational Burden Panels. These new panels collectively create a comprehensive profile of TMB and MSI status by “achieving high analytical sensitivity, with lower false and negative rates, while still maintaining >95% correlation with whole exome datasets.” The QIAseq TMB Panel covers 486 genes and can be boosted to add 27 MSI markers, with sample preparation coming from FFPE of solid tumors, patient’s plasma/serum, fresh or frozen tissue, or from cell lines in a research setting. This assay has been shown to correlate well with WES and established TMB panels. It uses enzymatic fragmentation for easy workflow, employs robust analysis modules, and incorporates unique molecular indices to correct for polymerase chain reaction (PCR) and sequencing errors (Qiagen, 2024).
TruSight Oncology 500
Illumina offers the TruSight Oncology 500 (formerly Illumina TSO500), a NGS assay that enables in house CGP of tumor samples, supports the identification of relevant DNA and RNA variants implicated in various solid tumor types, and accurately measure current immune-oncology biomarkers: MSI and TMB. This assay assesses both DNA (523 genes) and RNA (55 genes) for assessment of all variant types in addition to MSI and TMB. The TruSight Oncology 500 assay is intended for multiple solid tumor types and detects CNVs, gene fusions, insertions-deletions, single nucleotide variants, and transcript variants. It provides stealth-oncology biomarker coverage, including biomarkers of TMB and MSI, and is inclusive of human leukocyte antigen (HLA) regions, DNA polymerase epsilon catalytic subunit isoform b (POLE1), and DNA polymerase delta catalytic subunit (POLD1) (Illumina, 2024).
Analytical Validity
Wu et al. (2019) noted that targeted NGS panels often use correlation (R2) between panel- and WES- based TMB to validate TMB estimation, which can be distorted by cases where a tumor within a certain cancer type has a relatively ultra-high TMB. Thus, the authors proposed using accuracy (the proportion of cases correctly identified as TMB-high or TMB-low using panel based TMB) as a more robust indicator of panel performance, suggesting that accuracy and cancer type individualization are key factors to consider when designing panels for TMB estimation. The authors examined the TMB estimation from five available NGS panels for TMB determination (F1CDx, MSK-IMPACT, Illumina TSO500 [now TruSight Oncology 500], Oncomine TML, QIAseq TMB) to compare correlation to accuracy. F1CDx seemed to accurately assess TMB (R2≥0.75) in at least 24 out of 33 cancer types. However, when the cutoff point for high TMB was defined as the top 20% in each cancer type, the accuracy within these 24 cancer types ranged largely (56-99%), with only seven cancer types having satisfactory accuracy. These results indicate that F1CDx-based TMB estimation is only reliable in certain cancer types (cervical squamous cell carcinoma and endocervical adenocarcinoma, colon adenocarcinoma, head and neck squamous cell carcinoma, lung adenocarcinoma, skin cutaneous melanoma, stomach adenocarcinoma, and uterine corpus endometrial carcinoma). In the other 17 cancer types, correlation overestimated TMB status misclassifying a considerable number of TMB-low patients as TMB-high. This overestimation by correlation occurs because correlation is vulnerable to distortion by the common presence of cases with relatively ultra-high TMB within each cancer type. When removing cases with these relatively ultra-high TMB (top five percent within a particular cancer type), a dramatic decline in correlation between F1CDx- and WES-based TMB was observed in half of the 24 cancer types. In contrast, removing the top five percent of cases in each cancer type did not, in general, affect the accuracy within these 24 cancer types. Accuracy was also superior to correlation in the other four NGS panels examined. The authors conclude that the five available NGS panels can assess TMB accurately only in the seven cancer types aforementioned and note that for relatively ultra-high TMB, correlation is an unreliable evaluation of panel based TMB estimation performance in most cancer types. Instead, they find that accuracy is a superior index in this situation. They conclude that individualized panels by cancer type might be a better strategy to guarantee the most robust TMB estimation and provide better power in detecting the predictive function of TMB in these cancer types (Wu et al., 2019).
J. Willis et al. (2019) aimed to evaluate the validity of cell-free (cf) DNA to detect microsatellite instability (MSI). The authors validated the Guardant360 MSI detection with 1145 cf-DNA samples and further investigated the landscape of cf-DNA based MSI across 28459 plasma samples. Circulating tumor (ct, used as a proxy for cf-DNA) MSI assessment was compared to current standard of care tissue testing (a combination of immunohistochemistry, PCR, and NGS). The study included 949 patients that were considered unique and evaluable. The ctDNA method detected MSI-high values (82% accuracy) in 71 of 82 patients. A total of 863 of 867 patients reported as MSI-stable (99.5% accuracy) for a total of 98.4% accuracy amongst all MSI-reported groups. The positive predictive value was found to be 95%. Finally, the authors examined the clinical outcomes of 16 patients with MSI-H gastric cancer that were treated with immunotherapy. The authors found that 10 of 16 patients achieved “complete or partial remission with sustained clinical benefit” after being treated with immunotherapy (J. Willis et al., 2019).
Georgiadis et al. (2019) aimed to validate a noninvasive approach for detection of MSI and TMB. The authors used “a hybrid-capture–based 98-kb pan-cancer gene panel, including targeted microsatellite regions” as well as a “novel peak-finding algorithm…established to identify rare MSI frameshift alleles in cell-free DNA.” The authors evaluated 163 healthy patients with this assay, identifying a specificity of 99.4% (one false positive, which was considered a lower bound by the authors). The authors also evaluated 23 MSI-H cases (from patients with metastatic cancers) and six MSI-stable cases with the algorithm. Using cf-DNA, the algorithm detected MSI in 18 of the 23 MSI-H samples (78.3%) and correctly identified all six of the MSI-stable cases. Further, this approach was also applied to TMB (using a cutoff of 51 mutations/Mbp), of which the algorithm correctly detected 10 of the 15 samples identified as TMB-high (67%) (Georgiadis et al., 2019).
Woodhouse et al. (2020) evaluated the analytical performance of FoundationOne Liquid CDx assay to detect genomic alterations in cancer patients. The assay was evaluated across more than 30 different cancer types in over 300 genes and greater than 30,000 gene variants. “Results demonstrated a 95% limit of detection of 0.40% variant allele fraction for select substitutions and insertions/deletions, 0.37% variant allele fraction for select rearrangements, 21.7% tumor fraction (Marabelle, Le, et al.) for copy number amplifications, and 30.4% TF for copy number losses. The false positive variant rate was 0.013% (approximately one in 8,000). Reproducibility of variant calling was 99.59%” (Woodhouse et al., 2020). In comparison to in situ hybridization and immunohistochemistry, FoundationOne had an overall 96.3% positive percent agreement and >99.9% negative percent agreement. “These study results demonstrate that FoundationOne Liquid CDx accurately and reproducibly detects the major types of genomic alterations in addition to complex biomarkers such as microsatellite instability, blood tumor mutational burden, and tumor fraction” (Woodhouse et al., 2020).
Using data from the phase III MYSTIC trial, Si et al. (2021) validated the use of blood TMB (bTMB) using circulating cell-free tumor DNA (ctDNA) through comparisons with TMB measurements from tumor tissue (tTMB). Patients with metastatic NSCLC were treated either with first-line durvalumab (anti–PD-L1 antibody) ± tremelimumab (anticytotoxic T-lymphocyte-associated antigen-4 antibody) or chemotherapy, then bTMB and tTMB were evaluated using the GuardantOMNI and the FoundationOne CDx assay, respectively. To identify the optimal bTMB cutoff, the authors used a Cox proportional hazards model and minimal p value cross-validation. In the majority of patients in the MYSTIC study, somatic mutations were detected in ctDNA extracted from plasma samples, which allowed for the subsequent calculation of bTMB. The authors found that the success rate for obtaining valid TMB scores was higher for bTMB (81%) than for tTMB (63%) and that with minimal p value cross-validation analysis, they suggest a bTMB ≥20 mutations per megabase (mut/Mb) as the optimal cutoff for clinical benefit with durvalumab + tremelimumab. Overall, their study demonstrated that use of bTMB from plasma samples was feasible and provided accurate and reproducible TMB detection using the GuardantOMNI ctDNA platform. They also state that using “the new bTMB algorithm and an optimal bTMB cutoff of ≥20 mut/Mb, high bTMB was predictive of clinical benefit with durvalumab + tremelimumab versus chemotherapy” (Si et al., 2021).
Clinical Utility and Validity
Overman et al. (2017) examined MSI-H/dMMR colorectal cancer individual response to nivolumab, a PD-1 ICI. In this phase two trial, the authors assessed adult individuals with histologically confirmed recurrent or mCRC locally assessed as dMMR/MSI-H who had progressed either while still on or after, or who had been intolerant of, at least one prior line of treatment, including fluoropyrimidine and oxaliplatin or irinotecan. The primary endpoint of this study was investigator-assessed ORR. This study ran from March 12, 2014 to March 16, 2016 and included 74 individuals, most of whom (54.1%), had received more than three prior therapies. Twelve months following initiation of nivolumab treatment, 31.1% (23) of the individuals had an investigator-assessed ORR, which 68.9% of the individuals had disease control for more than 12 weeks. The median duration of response was not reached, as all responders were alive and eight of them had responses greater than 12 months. Twenty-three of the individuals died during the study, though none of the deaths were considered treatment-related. The authors concluded that nivolumab provided durable responses and disease control in dMMR/MSI-H patients that had previously received other forms of treatment for their colorectal cancer and they also note that nivolumab is a relevant treatment option for these patients (Overman et al., 2017).
Hellmann, Ciuleanu, et al. (2018) conducted an open-label, multipart, phase three trial to examine progression-free survival in non-small-cell lung cancer patients with a high TMB (TMB-H) that were being treated with nivolumab plus ipilimumab or with chemotherapy. TMB was determined by the FoundationOne CDx™ assay. Here, they found that progression-free survival in TMB-H patients was significantly longer with nivolumab plus ipilimumab (42.6%) compared with chemotherapy (13.2%). This discovery was found to be accurate irrespective of PD-L1 expression level and validated that a treatment of nivolumab and ipilimumab for NSCLC elicited a favorable outcome for select patients, as well as determining that tumor mutational burden was an important biomarker for patient selection when it comes to drug therapy (Hellmann, Ciuleanu, et al., 2018).
To examine the association between TMB and clinical response to ICIs in select cancer types, Samstein et al. (2019) analyzed the clinical and genomic data of 1,662 advanced cancer patients treated with ICI compared to 5,371 patients not treated with ICI. Tumors from these patients underwent targeted NGS with MSK-IMPACT. The authors found that among all patients, higher somatic TMB (highest 20% in each cancer type, consistent with many MSI-high colorectal tumors receiving ICI treatment) was associated with better overall survival. However, the TMB cutoffs associated with improved survival varied markedly between the different cancer types studied, suggesting that while TMB is associated with improved survival in patients receiving ICI across a variety of cancer types, there may not be a single, universal definition of high TMB (Samstein et al., 2019).
Ready et al. (2019) conducted an open-label phase II trial to evaluate the efficacy and safety of nivolumab plus low-dose ipilimumab as first-line treatment of advanced/metastatic NSCLC and assessed the treatment efficacy associated with PD-L1 expression and TMB. For this trial, “the primary end point was objective response rate (ORR) in patients with 1% or more and less than 1% tumor PD-L1 expression. Efficacy on the basis of TMB (FoundationOne CDx™ assay) was a secondary end point.” They found that regardless of PD-L1 expression, ORRs were higher in TMB-H patients versus patients with tumors below the threshold for TMB-H status and progression-free survival was longer in patients with TMB-H. They concluded that “nivolumab plus low-dose ipilimumab was effective and tolerable as a first-line treatment of advanced/metastatic NSCLC” and a high TMB status was associated with both improved response and prolonged progression-free survival irrespective of PD-L1 status. This identifies TMB-H as a potentially relevant cutoff in the assessment of TMB as a biomarker for first-line nivolumab plus ipilimumab (Ready et al., 2019).
A comprehensive study by Singal et al. (2019) examined the electronic health records of 4064 individuals with NSCLC from 275 different oncology practices to explore the associations between tumor genomics and patient characteristics with clinical outcome. The authors note that 21.4% of these individuals had a mutation in EGFR, ALK, or ROS1 and that patients with driver mutations who received targeted therapies had significantly improved overall survival times than individuals who did not receive targeted therapies (median of 18.6 versus 11.4 months, respectively). Moreover, a TMB of 20 or higher was associated with improved overall survival for patients on PD-L1-targeted therapy compared to patients with a TMB less than 20. TMBs measure the quality of a mutation in a tumor, suggesting whether a patient will benefit from immunology-based cancer therapies or not. The authors concluded that similar associations from previous research were replicated “between clinical and genomic characteristics, between driver mutations and response to targeted therapy, and between TMB and response to immunotherapy” (Singal et al., 2019).
Yarchoan et al. (2019) examined the relationship between the expression of PD-L1, a widely used biomarker for selecting patients for ICI, and TMB across the entire spectrum of ICI-responsive human cancers. They sought to use these predictive biomarkers to provide a broad definition to the immunologic subtypes of cancers and identify opportunities for the development of therapeutics. Their clinical cohort contained 9,887 unique clinical samples with paired CGP and PD-L1 expression obtained during standard clinical care. They found that 15.2% of samples had a positive expression of PD-L1 and that the highest occurrence of PD-L1 positivity occurred in thymic cancer and diffuse large B cell lymphoma, while there were no adenoid cystic or appendiceal samples with positive PD-L1. A total 3.6% of sample specimens were identified as having a high PD-L1 positivity (defined as greater than 50% of tumor cells staining positive). In addition, the median tumor mutational burden for all specimens was 3.48 mutations per megabase. One group of sampled tumors (16.4% of the cancers) had a TMB of greater than 10 mutations per megabase and 7.3% of tumors analyzed had a TMB of more than 20 mutations per megabase, indicating that the overall TMB profiles were comparable to prior cancer studies of a similar nature (including whole-exome studies) that estimated TMB biomarkers in solid tumors. This study found a relative independence of PD-L1 expression and TMB, and that the biomarkers were important indicators of patient response to ICI therapy. The study also defined which biomarkers may help identify future therapies through their ability to segment and profile different tumor types. In conclusion, they found that PD-L1 and TMB status may help determine whether the use of ICI therapy is indicated as well as pinpoint tumor types that would demonstrate a beneficial response to immune checkpoint inhibitor therapy (Yarchoan et al., 2019).
Alborelli et al. (2020) investigated the predictive power of TMB for patients treated with ICIs. Seventy-six NSCLC patients treated with ICIs were included and TMB was evaluated with the Oncomine™ Tumor Mutational Load (TML) sequencing assay. Patients were separated into cohorts of “durable clinical benefit” (DCB) or “no durable benefit” (NDB). TMB was found to be higher in the DCB cohort (median TMB of 8.5 mutations / Mb compared to six mutations / Mb in NDB). Sixty-four percent of patients in the highest one third of TMB were responders, compared to 33% and 29% in the middle and lowest thirds, respectively. TMB-H patients were also found to have higher progression-free survival and overall survival. Overall, the authors concluded that the TML panel was an effective tool to stratify patients for ICI treatment and suggested that “a combination of biomarkers might maximize the predictive precision for patient stratification.” Further, the authors remarked that their data “supports TMB evaluation through targeted NGS in NSCLC patient samples as a tool to predict response to ICI therapy” (Alborelli et al., 2020).
Marabelle, Fakih, et al. (2020) prospectively explored the association of a high tumor mutational burden with outcomes in ten tumor-type-specific cohorts from the phase two KEYNOTE-158 study. The study assessed use of the anti–PD-1 monoclonal antibody pembrolizumab in patients with select, previously treated, advanced solid tumors. Patients for this study were from 81 academic facilities across 21 countries in Africa, the Americas, Asia, and Europe, were over 18 years of age, and had an established tumor of anal, biliary, cervical, endometrial, mesothelioma, neuroendocrine, salivary, small-cell lung, thyroid, or vulvar origin. In addition, patients showed continued tumor progression or an inability to tolerate standard therapy and continued measurable disease (based on the Response Evaluation Criteria in Solid Tumors). The treatment course for this study was pembrolizumab 200 mg intravenously every three weeks for up to thirty-five cycles, with tissue TMB assessed in formalin-fixed, paraffin-embedded tumor samples using the FoundationOne CDx assay. The authors report that 13% of the participants showed a high number of tumor mutations (≥ 10 mutations per megabase) while 87% of the participants had a TMB status lower than this threshold. From this study, they found that a high TMB status characterized a small group of patients who might have a beneficial response to pembrolizumab therapy; those with previously treated recurrent or advanced solid tumors. As such, tissue TMB may be both a “novel and useful predictive biomarker” for therapy indications (Marabelle, Fakih, et al., 2020).
Data from the KEYNOTE-158 study was also analyzed by to examine the efficacy of pembrolizumab as an ICI treatment for MSI-H.dMMR cancer. The authors examined the efficacy of pembrolizumab in patients with previously treated, advanced noncolorectal MSI-H/DMMR cancer from the phase II KEYNOTE-158 study. In the 233 enrolled patients, 27 tumor types were represented (endometrial, gastric, cholangiocarcinoma, and pancreatic cancers were the most common). ORR was 34.3%, median PFS was 4.1 months, and median OS was 23.5 months. Adverse, treatment-related events occurred in 64.8% of patients (151) and 14.6% (34) patients had grade three to five treatment-related fatal adverse events. They report that pembrolizumab is clinically beneficial in patients with previously treated, unresectable or metastatic MSI-H/dMMR noncolorectal cancer and that toxicity of pembrolizumab monotherapy in these patients was consistent with previous data (Marabelle, Le, et al., 2020).
A study by Stahler et al. (2020) explored the ORR, PFS, and OS in the FIRE-3 clinical trial on metastatic colorectal cancer patients who were identified by the FoundationOne CDx panel to have SNVs, copy number alterations, TMB-H, or MSI-H tumors. Of the 752 patients in the FIRE-3 trial, 373 provided material for this specific analysis. They found that MSI-H (30%) and TMB-H (17.3%) tumors were enriched by FBXW7 mutations a frequent SNV found in the analysis. The authors found that “RAS, BRAF V600E and SMAD4 mutations were identified as poor prognostic biomarkers in patients of the FIRE-3 trial, whereas improved outcome was observed for BRAF non-V600E mutation. SMAD4 mutation might provide predictive relevance for cetuximab efficacy. MSI-H tumours showed numerically lower ORR, OS and PFS” (Stahler et al., 2020).
While ICIs are highly effective in patients with MSI/dMMR metastatic colorectal cancer (mCRC), Cohen et al. (2020) predicted that the Response Evaluation Criteria in Solid Tumors (RECIST) 1.1 criteria was underestimating the response to ICIs due to the pseudoprogression phenomenon. Using data from the GERCOR NIPICOL phase II study, the authors aimed to evaluate the frequency of pseudoprogressions in patients with MSI/dMMR mCRC that were treated with nivolumab and ipilimumab. Nivolumab was given to MSI/dMMR mCRC patients previously treated with fluoropyrimidines, oxaliplatin, and irinotecan with/without targeted therapies. The primary endpoint of the study was disease control rate at 12 weeks according to RECIST 1.1 and iRECIST by central review. Fifty-seven patients were included in the study, with 48% having received ≥ three prior lines of chemotherapy, 18% with BRAF mutations, and 56% with Lynch syndrome-related cancer. Of these 57 patients, seven discontinued due to adverse events and one died due to a treatment-related adverse events (TRAE). The authors found that at 12 weeks, the disease control rate was 86% with RECIST 1.1 and 87.7% with iRECIST and two pseudoprogressions were observed (one at week 6, one at week 36). These two pseudoprogressions represented 18% of patients with disease progression per RECIST 1.1 criteria. The 12-month PFS rate was 72.9% with RECIST 1.1 and 76.5% with iRECIST and the 12-month OS rate was 84%. The ORR was 59.7% with both criteria. The authors concluded that pseudoprogression is rare in patients with MSI/dMMR mCRC who are treated with nivolumab and ipilimumab and that this combined ICI therapy confirms impressive disease control rate and survival outcomes in these MSI/dMMR patients (Cohen et al., 2020).
Andre et al. (2020) examined the efficacy of first-line pembrolizumab therapy as compared to first-line chemotherapy therapy for dMMR/MSI-H advanced or metastatic colorectal cancer. In this phase three, open-label clinical trial, 307 patients with metastatic MSI/dMMR colorectal cancer who had not received prior cancer treatment were randomly assigned to pembrolizumab or chemotherapy as a primary treatment and PFS and OS were assessed. Patients that received chemotherapy were able to cross over to pembrolizumab therapy if their disease progressed. The authors found that pembrolizumab treatment in MSI-H/dMMR colorectal cancer patients was superior to chemotherapy with respect to PFS. At the time of cutoff, 56 members of the pembrolizumab group and 69 chemotherapy group patients had died. At the time of the report, OS data were still evolving (they report that 66% of required events had occurred). An overall response (complete or partial) was observed in 43.8% of pembrolizumab patients as compared to only 33.1% of chemotherapy patients. Among those patients with an overall response, 83% of the pembrolizumab group, as compared to just 35% of the chemotherapy group, had an ongoing response at 24 months. TRAEs occurred in just 22% of pembrolizumab patients as compared to 66% of chemotherapy patients (including one death). The authors conclude that pembrolizumab is superior to chemotherapy as a first-line therapy in MSI-H/dMMR metastatic colorectal cancer, with fewer TRAE (Andre et al., 2020).
Wei et al. (2022) evaluated Illumina’s TruSight Oncology 500 (TSO500) Assay for its ability to assess tumor mutational burden and the clinical utility of predicting response to pembrolizumab. Using samples collected from approximately 300 patients with “at least twelve different types of advanced solid tumors enrolled in eight clinical trials of pembrolizumab monotherapy,” and using FoundationOneCDx and WES as reference methods for TMB, the researchers found that the TSO500 was able to properly classify the status of high tumor mutational burden, a reproducible finding with subsequent use of the assay. The researchers also found that “similar to the validated and FDA-approved TMB cut-point of 10 mut/Mb assessed by FoundationOneCDx, the TSO500 TMB cut-point of 10 mut/Mb is predictive of response to pembrolizumab monotherapy.” Ensuring the comparability of this assay to current technologies in the necessary settings may increase access to immunotherapy for patients with solid tumors and TMB-H (Wei et al., 2022).
Aggarwal et al. (2023) completed a clinical validation study of a TMB biomarker in the aim of assessing the association between TMB and clinical outcomes. The study included 674 patients with solid tumors from eight different cancer types. Patients were collected from 300 cancer sites between 2018 and 2022. All patients were sequenced with Tempus xT and treated with ICIs. The authors studied the association between the TMB binary category, high or low, and overall survival. Overall, “high TMB (TMB-H) cancers (206 patients [30.6%]) were significantly associated with longer OS than low TMB (TMB-L) cancers.” Additionally, “an overall survival benefit was seen regardless of the type of ICI used.” The authors concluded that “TMB-H cancers were significantly associated with improved clinical outcomes compared with TMB-L cancers” (Aggarwal et al., 2023).
National Comprehensive Cancer Network (NCCN)
The NCCN includes TMB and MSI testing in several of their guidelines for different types of cancers. Across these guidelines, TMB-high (TMB-H) is defined as tumors with a TMB value of ≥10 mutations/megabase.
The NCCN has clinical practice guidelines for NSCLC. In 2020 the NCCN deleted TMB as an emerging immune biomarker based on clinical trial data and other issues, though was previously considered potentially useful for selecting patients to be treated with nivolumab with or without ipilimumab. However, recent data showed that overall survival was improved with nivolumab/ipilimumab regardless of TMB status. They conclude that PD-L1 expression level is a more useful immune biomarker than TMB for deciding how to prescribe immunotherapy in NSCLC patients in some instances. For example, “Testing for PD-L1 expression levels is recommended before prescribing pembrolizumab monotherapy”, but “is not required for prescribing subsequent therapy with atezolizumab or nivolumab”, though it may still provide useful information. Moreover, the NCCN guidelines “do not recommend measurement of TMB levels before deciding whether to use nivolumab plus ipilmumab regimens or to use other ICIs, such as pembrolizumab” (NCCN, 2024l).
The NCCN clinical practice guidelines for cervical cancer advise that for stage IVB or distant metastases, “comprehensive molecular profiling as determined by an FDA-approved assay, or a validated test performed in a CLIA-certified laboratory” should be considered. They repeat this under their section for principles of pathology for squamous cell carcinoma, adenocarcinoma, and adenosquamous carcinoma, and add on that they recommend “PD-L1 testing for patients with recurrent, progressive, or metastatic disease”. The NCCN also recommend “mismatch repair (MMR)/microsatellite instability (MSI) testing for patients with recurrent, progressive, or metastatic cervical carcinoma” and/or “NTRK gene fusion testing for patients with cervical sarcoma”. For systemic cervical cancer therapy, the NCCN reports that pembrolizumab is a preferred regimen as a second-line or subsequent therapy for TMB-H, PD-L1–positive, or MSI-H/dMMR tumors that are recurrent or metastatic squamous cell carcinoma, adenocarcinoma, or adenosquamous carcinoma (NCCN, 2024g).
The NCCN clinical practice guidelines for breast cancer note that pembrolizumab is indicated for the treatment of patients with either unresectable or metastatic, MSI-high or mismatch repair-deficient (dMMR) solid tumors, or TMB-H breast tumors that have progressed following prior treatment and have no satisfactory alternative treatment options. TMB-H status is detected via NGS and MSI is detected via immunohistochemistry, NGS, or PCR. The NCCN finds using TMB and MSI to guide pembrolizumab treatment is associated with category 2A evidence (based upon lower-level evidence, there is uniform NCCN consensus that the intervention is appropriate). Dostarlimab-gxly is indicated for adult patients with MSI-H/dMMR unresectable or metastatic tumors and (category 2A) solid tumors or for TMB-H tumors that have progressed following prior treatment and with no satisfactory alternative treatment options. (NCCN, 2024f).
In their clinical practice guidelines for uterine neoplasms, the NCCN recommends that one should “Consider tumor mutational burden (TMB) testing through an FDA-approved assay, or a validated test performed in a CLIA-certified laboratory” as one of their principles of molecular analysis. The NCCN also notes the use of pembrolizumab for TMB-H or MSI-H/deficient DNA mismatch repair (dMMR) tumors as “useful in certain circumstances” as a biomarker-directed systemic therapy for uterine sarcoma and endometrial carcinoma. There, the NCCN also “recommends TMB-H testing if not previously done. Pembrolizumab is indicated for patients with unresectable or metastatic tumors with TMB-H [≥10 mutations/ megabase (mut/Mb)], as determined by an FDA-approved assay, or a validated test performed in a CLIA-certified laboratory, whose disease has progressed following prior treatment and who have no satisfactory alternative treatment options.” In the molecular analysis of sarcoma, it is recommended that “Comprehensive genomic profiling in setting of metastatic disease as determined by an FDA-approved assay, or a validated test performed in a CLIA-certified laboratory, is informative for predicting rare pan-tumor targeted therapy opportunities and should include at least NTRK, MSI, and TMB”(NCCN, 2024t).
The NCCN guidelines for colon cancer state that “determination of tumor gene status for KRAS/NRAS and BRAF mutations, as well as HER2 amplifications and MSI/MMR status (if not previously done), are recommended for patients with mCRC.” They note that “Testing may be carried out for individual genes or as part of an NGS panel, although no specific methodology is recommended.” However, NGS panels do have an advantage in that they are “able to pick up rare and actionable genetic alterations, such as neurotrophic tyrosine receptor kinase (NTRK) and rearranged during transfection (RET) fusions and may be carried out using either a tissue or blood-based (eg, liquid) biopsy” (NCCN, 2024h). The NCCN advises that “As with other conditions in which stage IV disease is diagnosed, a tumor analysis (metastases or original primary) for KRAS/NRAS and BRAF mutations and HER2 amplifications, as well as MSI/MMR testing if not previously done, should be performed to define whether targeted therapies can be considered among the potential options”. In pedunculated or sessile colon polyps (adenoma) with invasive cancer, MSI/MMR testing can be part of the workup. Colon cancer appropriate for resection (non-metastatic) will “require a complete staging workup, including biopsy, pathologic tissue review, total colonoscopy, CBC, chemistry profile, CEA determination, and baseline CT scans of the chest, abdomen, and pelvis”, while “Testing for MMR/MSI should be done at diagnosis to help with detection of Lynch syndrome and to inform treatment decision-making.” When evaluating the recurrence of documented metachronous metastases by CT, MRI, and/or biopsy, dMMR and MSI-H can be considered endpoints for workups (NCCN, 2024h).
In the NCCN clinical practice guidelines for head and neck cancer note that in cancer that occurs in the oral-cavity, image-guided (ultrasound [US] or CT) needle biopsy of cystic neck nodes may offer better diagnostic yield than fine-needle aspiration (FNA) by palpation alone for initial diagnosis. For unresectable or metastatic disease where there is a plan for systemic therapy, a core biopsy would allow for ancillary immune-genomic testing.” It is a requirement for cancer that occurs in the oropharynx be tested for tumor human papillomavirus (HPV) by p16 immunohistochemistry (IHC). Biopsy of primary site or FNA of the neck is recommended (NCCN, 2024w). For cancer that occurs in the Nasopharynx with nonkeratinizing or undifferentiated histology, it is recommended to test for EBV in tumor and blood. Common means for detecting EBV in pathologic specimens include ISH for EBV-encoded RNA or immunohistochemical staining for latent membrane protein (LMP). The EBV DNA load within the serum or plasma may be quantified using PCR targeting genomic sequences of the EBV DNA such as BamHI-W, Epstein-Barr virus nuclear antigen (EBNA), or LMP; these tests vary in their sensitivity. The EBV DNA load may reflect prognosis and change in response to therapy. Cancer that occurs in the salivary glands, “use NGS profiling and other appropriate biomarker testing to check status of androgen receptor (AR), HER2, NTRK, HRAS, PIK3CA, and tumor mutational burden (TMB) prior to treatment”(NCCN, 2024w).
In the NCCN clinical practice guidelines for esophageal and EGJ cancers, the panel outlines their principles of pathologic review and biomarker testing. There, they assert that “Immunohistochemistry/in situ hybridization/targeted PCR should be considered first for the identification of biomarkers, followed by NGS testing. If limited tissue is available, or the patient is unable to undergo a traditional biopsy, sequential testing of single biomarkers/limited molecular diagnostic panels will exhaust the sample. In these scenarios, or at the discretion of the treating physician, comprehensive genomic profiling via a validated NGS assay performed in a CLIA-approved laboratory should be considered”. Their list of targeted biomarkers include HER2 overexpression/amplification, PD-L1 expression by immunohistochemistry, microsatellite instability, tumor mutational burden, NTRK gene fusion, RET gene fusion, and BRAF V600E mutation (NCCN, 2024i). Furthermore, during workup for esophageal and esophagogastric junction cancers, “Universal testing for microsatellite instability (MSI) by PCR/next-generation sequencing (NGS) or MMR by IHC is recommended in all newly diagnosed patients”. They also recommend “Programmed death ligand 1 (PD-L1) testing if advanced/ metastatic disease is documented/suspected”, “HER2 testing if metastatic adenocarcinoma is documented/ suspected”, and that “NGS should be considered”. Universal testing for MSI by PCR/NGS or MMR by IHC is recommended in all newly diagnosed patients for biopsy specimen types, endoscopic resection, and esophagogastrectomy, without prior chemoradiation (if MSI and MMR has not been previously performed) (NCCN, 2024i). In their assessment of overexpression or amplification of HER2 in esophageal and EGJ cancers, the NCCN recommends that “The use of IHC/ISH should be considered first, followed by NGS testing as appropriate. Repeat biomarker testing may be considered at clinical or radiologic progression for patients with advanced/metastatic esophageal/EGJ adenocarcinoma” (NCCN, 2024i).
The NCCN clinical practice guidelines for occult primary (cancer of unknown primary [CUP]) note that “MSI/mismatch repair (MMR) testing is indicated for patients with CUP; however, it should be noted that the population of patients with MSI-high/MMR-deficient (MSI-H/dMMR) occult primary tumors is generally low.” Moreover, they remark that determination of TMB by a validated and/or FDA-approved assay is a category 2B recommendation (2B: based on lower-level evidence, there is NCCN consensus that the intervention is appropriate) and a biopsy from a suspected metastatic malignancy should undergo TMB testing during the workup of the tumor. The NCCN also asserts that during workup “Gene sequencing to predict tissue of origin is not recommended” and “Molecular profiling of tumor tissue using next-generation sequencing (NGS) (or other technique to identify gene fusions) can be considered after an initial determination of histology has been made”. Again, the NCCN cautions that “The population of patients with microsatellite instability-high/mismatch repair-deficient (MSI-H/dMMR) occult primary tumors is low. Use immunohistochemistry (IHC) for MMR or polymerase chain reaction (PCR) for MSI, which are different assays measuring the same biological effect” (NCCN, 2024m). For occult primary tumors, dostarlimab-gxly is recommended as “useful in certain circumstances” only in dMMR/MSI-H tumors, while pembrolizumab may be extended to tumors with either TMB-H or dMMR/MSI-H (NCCN, 2024m).
In the NCCN clinical practice guidelines for gastric cancer, universal testing, no matter the specimen type, for MSI by PCR/NGS or MMR by IHC is recommended in all newly diagnosed patients in the biopsy stage, endoscopic mucosal resection, gastrectomy without prior chemoradiation (if not previously performed). In the palliative management of unresectable locally advanced, locally recurrent, or metastatic disease, “Perform HER2, programmed death ligand 1 (PD-L1), and microsatellite testing (if not done previously) if metastatic cancer is documented or suspected” and “NGS may be considered via a validated assay”. Tumors should be interpreted as MSI-stable, MSI-low (1-29% of markers exhibit instability, one of the five NCI or mononucleotide markers exhibits instability), or MSI-H (≥30% of the markers exhibit instability, two or more of the five NCI or mononucleotide markers exhibit instability). They also state that “If limited tissue is available, or the patient is unable to undergo a traditional biopsy, sequential testing of single biomarkers/limited molecular diagnostic panels will exhaust the sample. In these scenarios, or at the discretion of the treating physician, comprehensive genomic profiling via a validated NGS assay performed in a CLIA-approved laboratory should be considered.” Their list of targeted biomarkers includes HER2 overexpression/amplification, PD-L1 expression by immunohistochemistry, microsatellite instability, tumor mutational burden, NTRK gene fusion, RET gene fusion, and BRAF V600E mutation (NCCN, 2024j).
The NCCN clinical practice guidelines for hepatocellular carcinoma state that there is “no established role for microsatellite instability (MSI), mismatch repair (MMR), tumor mutational burden (TMB), or PD-L1 testing in [hepatocellular carcinoma] at this time”. Immune checkpoint inhibition has shown clinical benefit leading to regulatory approvals in patients with HCC without selection for MSI, MMR, TMB, or PD-L1 status. Regarding therapies, the panel recommends nivolumab + ipilimumab (category 2B) as a first-line systemic therapy that is useful in certain circumstances and a subsequent-line systemic therapy if there is disease progression for individuals with TMB-H tumors. The NCCN also dostarlimab-gxly (category 2B) as a subsequent-line systemic therapy if there is disease progression for individuals with MSI-H/dMMR tumors (NCCN, 2024x).
The NCCN clinical practice guidelines for biliary tract cancers (BTCs) recommend germline testing and/or referral to a genetic councilor for patients with dMMR/MSI-H tumors or a family history suggestive of BRCA1/2 mutation” when presenting during intraoperative staging biopsy following an incidental finding of suspicious mass during surgery. Germline testing and/or referral to genetic counseling for patients with dMMR/MSI-H tumors or a family history suggestive of BRCA1/2 mutations should also be extended to situations where hepatobiliary surgery expertise is unavailable and postoperative workup is deemed unresectable, incidental finding on pathologic review with T1b or greater and/or T1a with positive margins deemed unresectable, unresectable mass on imaging, jaundice, metastatic disease, or isolated intrahepatic mass with unresectable or metastatic disease biopsy. The NCCN recommends that “Comprehensive molecular profiling is recommended for patients with unresectable or metastatic BTC who are candidates for systemic therapy”. They also note that “Evidence remains insufficient for definitive recommendations regarding specific criteria to guide genetic risk assessment in hepatobiliary cancers or for universal germline testing in these tumors” and, as such, “In BTCs, genetic counseling referral and potential germline testing should be considered in patients with any of the following characteristics: young age at diagnosis; a strong personal or family history of cancer; no known risk factors for liver disease; or presence of mutations identified during tumor testing that are suspected to be possible germline alterations. For patients who do harbor a known germline mutation associated with a cancer predisposing syndrome (ie, Lynch syndrome or hereditary breast and ovarian cancer syndrome), there is currently insufficient evidence to support screening for biliary tract malignancies” (NCCN, 2024d). The NCCN asserts that “testing for MSI or MMR deficiency is recommended in patients with unresectable or metastatic gallbladder cancer, intrahepatic CCA, or extrahepatic CCA” and that “Testing for TMB is recommended for patients with unresectable or metastatic gallbladder cancer, intrahepatic CCA, or extrahepatic CCA based upon clinical benefit observed across advanced solid tumors.” They also outline the three possible tests to evaluate mismatch repair (MMR) protein deficiency or microsatellite status. First, an immunohistochemical staining for the MLH1, MSH2, MSH6, and PMS2 gene products will establish protein retention or loss, followed by NGS to determine if there are inactivating mutations in the MMR genes (MLH1, MSH2, MSH6, and PMS2). Lastly, “microsatellite repeats of tumor DNA are examined by polymerase chain reaction (PCR)” and caution that “Tumor mutational burden (TMB) can be tested with a clinically validated NGS panel but has inherent platform variation” (NCCN, 2024d). In addition, the NCCN provides a helpful graphic to show their recommendations for molecular testing in unresectable or BTCs, captured below:

(NCCN, 2024d).
The NCCN clinical practice guidelines for neuroendocrine and adrenal tumors recommends that TMB and MSI testing should be considered as part of the workup for adrenocortical carcinoma. In the evaluation of poorly differentiated or large or small cell carcinomas or unknown primary, “Tumor biomarkers such as microsatellite instability (MSI), MMR, and TMB testing (by an FDA-approved test) should be considered as they can aid in assessing targeted therapy options”. In well-differentiated grade three neuroendocrine tumors, “Tumor/somatic molecular profiling should be considered for patients with locoregional unresectable/metastatic disease who are candidates for anti-cancer therapy to identify actionable alterations. Testing on tumor tissue is preferred; however, cell-free DNA testing can be considered if tumor tissue testing is not feasible.” The NCCN notes that “patients with ACC may also consider MSI, MMR, and TMB (by an FDA-approved test) testing” because some analyses suggest that “3% of patients with ACC have Lynch syndrome” (NCCN, 2024k).
The NCCN clinical practice guidelines for vulvar cancer state that “Mismatch repair (MMR), microsatellite instability (MSI), programmed death ligand 1 (PD-L1), neurotrophic tyrosine receptor kinase (NTRK) gene fusion, and tumor mutational burden (TMB) testing may also be considered for treatment planning purposes in patients with recurrent, progressive, or metastatic disease” (NCCN, 2024u).
The NCCN clinical practice guidelines for ovarian cancer identify TMB testing as a clinically indicated tumor molecular analysis in addition to NGS for BRCA1/2 mutations and other somatic mutations. They also recommend using tumor molecular analysis “to identify potential benefit from targeted therapeutics that have tumor-specific or tumor-agnostic benefit including, but not limited to, HER2 status (by IHC), BRCA1/2, HRD status, microsatellite instability (MSI), mismatch repair (MMR), tumor mutational burden (TMB), BRAF, FRα (FOLR1), RET, and NTRK if prior testing did not include these markers” in patients with recurrent disease and rising CA-125, regardless of prior chemotherapy status if molecular testing was not done previously in epithelial ovarian cancer/fallopian tube cancer/primary peritoneal cancer. The NCCN guidelines also recommend tumor molecular testing with TMB and MSI if not previously done when following up or monitoring for the recurrence of low-grade serous carcinoma (NCCN, 2024n).
In the NCCN clinical practice guidelines for testicular cancer, the NCCN recommends TMB or MSI testing as a third-line therapy in nonseminoma if there is progression after high-dose chemotherapy or other third-line therapies. The guideline also notes that similar to many other cancers, pembrolizumab may be useful in certain circumstances as a third-line chemotherapy regimen for metastatic germ cell tumors when the tumor has a TMB-H or MSI-H status, regardless of prior high-dose chemotherapy status (NCCN, 2024r).
In the NCCN clinical practice guidelines for pancreatic adenocarcinoma, it is stated that “Tumor/somatic molecular profiling is recommended for patients with locally advanced/metastatic disease who are candidates for anti-cancer therapy to identify uncommon mutations. Consider specifically testing for potentially actionable somatic findings including, but not limited to: fusions (ALK, NRG1, NTRK, ROS1, FGFR2, and RET), mutations (BRAF, BRCA1/2, KRAS, and PALB2), amplifications (HER2), microsatellite instability (MSI), mismatch repair deficiency (dMMR), or tumor mutational burden (TMB) via an FDA-approved and/or validated next-generation sequencing (NGS)-based assay. RNA sequencing assays are preferred for detecting RNA fusions because gene fusions are better detected by RNA-based NGS. Testing on tumor tissue is preferred; however, cell-free DNA testing can be considered if tumor tissue testing is not feasible”. This is true for locally advanced disease where adenocarcinoma is confirmed, in metastatic disease when tumor tissue is available, and in tumors with recurrence after resection (NCCN, 2024b).
In the NCCN clinical practice guidelines for penile cancer, pembrolizumab is recommended as a subsequent-line systemic therapy for metastatic/recurrent disease in MSI-H/dMMR tumors that have progressed following prior treatment and that have no satisfactory alternative treatment options (NCCN, 2024c).
The NCCN clinical practice guidelines for prostate cancer recommend that in CRPC, tumor testing for microsatellite instability (MSI-H) or DMMR and HRRm should be considered when CRPC imaging studies are positive for metastases, if the testing has not been previously performed. It also recommends tumor testing for MSI-H or dMMR among patients with metastatic CRPC and “may be considered in patients with regional or castration-sensitive metastatic prostate cancer”, and that “TMB testing may be considered in patients with metastatic castration-resistant prostate cancer.” The NCCN also provides tumor testing recommendations based on risk groups. These recommendations include
- “Tumor testing for somatic homologous recombination gene mutations (eg, BRCA1, BRCA2, ATM, PALB2, FANCA, RAD51D, CHEK2, CDK12) can be considered in patients with regional (N1) prostate cancer and is recommended for those with metastatic disease.”
- “Tumor testing for MSI or dMMR can be considered in patients with regional or metastatic castration-naïve prostate cancer and is recommended in the metastatic CRPC setting.”
- “Tumor mutational burden (TMB) testing may be considered in patients with metastatic CRPC.”
- “Multigene molecular testing can be considered for patients with low-, intermediate-, and high-risk prostate cancer and life expectancy ≥10 years.”
- “The Decipher molecular assay is recommended to inform adjuvant treatment if adverse features are found post-radical prostatectomy, and can be considered as part of counseling for risk stratification in patients with PSA resistance/recurrence after radical prostatectomy” (NCCN, 2024o).
The NCCN advises that “If MSI testing is performed, testing using an NGS assay validated for prostate cancer is preferred” and states that “If MSI-H or dMMR is found, the patient should be referred for genetic counseling to assess for the possibility of Lynch syndrome” (NCCN, 2024o).
The NCCN clincal practice guidelines for rectal cancer state that universal MMR or MSI testing is recommended in all newly diagnosed patients with rectal cancer, noting that IHC for MMR and DNA analysis for MSI are different assays and measure different biological effects caused by deficient MMR function (NCCN, 2024p). The NCCN cautions that “The presence of a BRAF V600E mutation in the setting of MLH1 absence would preclude the diagnosis of Lynch syndrome (LS) in the vast majority of patients. However, approximately one percent of cancers with BRAF V600E mutations (and loss of MLH1) are LS. Caution should be exercised in excluding patients with a strong family history from germline screening in the case of BRAF V600E mutations.” The further explain that “One of two different initial tests can be performed on CRC specimens to identify individuals who might have Lynch syndrome: 1) immunohistochemical analysis for MMR protein expression, which is often diminished because of mutation; or 2) analysis for microsatellite instability (MSI), which results from MMR deficiency and is detected as changes in the length of repetitive DNA elements in tumor tissue caused by the insertion or deletion of repeated units”. They also remind clinicians that “MMR or MSI testing should be performed only in CLIA-approved laboratories” and that “testing for MSI may be accomplished by polymerase chain reaction (PCR) or a validated NGS panel, the latter especially in patients with metastatic disease who require genotyping of RAS and BRAF”. In the management of malignant polyps, “All patients with a malignant polyp should undergo MMR or MSI testing at diagnosis”. Similarly, “All patients with rectal cancer should undergo MMR or MSI testing at diagnosis to aid in the diagnosis of Lynch syndrome and for clinical trial availability, especially related to checkpoint inhibitors as neoadjuvant therapy” (NCCN, 2024p). MMR and MSI testing is also recommended as part of the initial workup in pedunculated polyps or sessile polyps (adenoma) with invasive cancer and in rectal cancers with and without suspected or proven distant metastases (if not previously done in the latter). Notably, during pathologic review, “It has not been established if molecular markers (other than MSI-H/dMMR) are useful in treatment determination (predictive markers) and prognosis” (NCCN, 2024p).
The NCCN clinical practice guidelines for small bowel adenocarcinoma recommend MSI testing as a workup when the tumor is in the duodenum or the jejunum/ileum, as well as in metastatic and/or recurrent tumors such as metastatic adenocarcinoma. Some general principles of pathologic review include the following:
- “Universal MMR or MSI testing is recommended in all newly diagnosed patients with SBA”
- “Patients with stage II dMMR/MSI-H may have improved survival compared to patients with proficient MMR (pMMR)/microsatellite stable (MSS); however, this has not been confirmed in the SBA population and is extrapolated from colorectal cancer data”
- “MMR or MSI testing should be performed only in Clinical Laboratory Improvement Amendments (CLIA)-certified laboratories”
- “Testing for MSI may be performed by validated next-generation sequencing (NGS) panels” (NCCN, 2024q).
The NCCN clinical practice guidelines for thyroid carcinoma suggest that treatment of structurally persistent/recurrent locoregional or distant metastatic disease not amenable to RAI therapy, “somatic testing to identify actionable mutations (including ALK, NTRK, BRAF, and RET gene fusions), mismatch repair deficiency (dMMR), microsatellite instability (MSI), and tumor mutational burden (TMB)” should be considered for advanced, progressive, or threatening disease. Wh en FNA or core biopsy finds that it is anaplastic thyroid carcinoma, molecular testing is recommended as diagnostic molecular testing for actionable mutations, which they propound as including “BRAF, NTRK, ALK, RET, MSI, dMMR, and tumor mutational burden. BRAF IHC testing is recommended due to faster turnaround compared to genetic testing.” This extends to the aggressive therapy approach for stage IVC (NCCN, 2024s).
The NCCN clinical practice guidelines for bone cancer recommend considering TMB or MSI testing as determined by a validated and/or FDA-approved assay to inform the use of treatment options in patients with metastatic chondrosarcoma, chordoma, Metastatic Ewing sarcoma, and metastatic osteosarcoma at presentation (NCCN, 2024e).
The NCCN clinical practice guidelines for ampullary adenocarcinoma suggest that if adenocarcinoma is found to be metastatic, then “Tumor/somatic molecular profiling is recommended for patients with locally advanced/metastatic disease who are candidates for anti-cancer therapy to identify uncommon mutations”, and recommend that clinicians “Consider specifically testing for potentially actionable somatic findings including, but not limited to: fusions (ALK, NRG1, NTRK, ROS1, FGFR2, and RET), mutations (BRAF, BRCA1/2, KRAS, and PALB2), amplifications (HER2), microsatellite instability (MSI), mismatch repair deficiency (dMMR), or tumor mutational burden (TMB) via an FDA-approved and/or validated next-generation sequencing (NGS)-based assay.” The NCCN panel also explains that “RNA sequencing assays are preferred for detecting RNA fusions because gene fusions are better detected by RNA-based NGS”; regardless, “Testing on tumor tissue is preferred; however, cell-free DNA testing can be considered if tumor tissue testing is not feasible” (NCCN, 2024a).
The NCCN clinical practice guidelines for anal carcinoma note that “microsatellite instability (MSI)/mismatch repair (MMR) testing is not required” during second-line treatment of metastatic anal cancer because “MSI is uncommon in anal cancer; [however], anal cancers may be responsive to PD-1/PD-L1 inhibitors because they often have high PD-L1 expression and/or a high tumor mutational load despite being microsatellite stable (MSS) ” (NCCN, 2024v).
The NCCN clinical practice guidelines for hepatocellular carcinoma note that “There is no established role for microsatellite instability (MSI), mismatch repair (MMR), tumor mutational burden (TMB), or programmed death ligand 1 (PD-L1) testing in HCC at this time. Immune checkpoint inhibition has shown clinical benefit leading to regulatory approvals in patients with HCC without selection for MSI, MMR, TMB, or PD-L1 status” (NCCN, 2024x).
The NCCN clinical practice guidelines for pediatric central nervous system cancers mention TMB in terms of palliative systemic therapy for recurrent or progressive disease, noting that “targeted therapy based on the molecular composition of the tumor is recommended for patients with good performance status” including “checkpoint blockade for high tumor mutational burden (TMB) or personal or family history of cMMRD; RAF and MEK inhibition for tumors with BRAF V600E mutation, and TRK inhibitors for tumors with NTRK gene fusion” (NCCN, 2024y).
The NCCN clinical practice guidelines for soft tissue sarcoma recommends pembrolizumab as useful in certain circumstances as a subsequent line of therapy for advanced/metastatic disease for MSI-H or dMMR tumors (regardless of soft tissue sarcoma sub-type). The NCCN also recommends pembrolizumab or nivolumab ± ipilimumab as useful in certain circumstances as a subsequent line of therapy for advanced/metastatic disease for TMB-H (≥10 mutations/megabase [mut/Mb]) regardless of soft tissue sarcoma sub-type (NCCN, 2024z).
European Society for Medical Oncology (ESMO)
The ESMO has published clinical recommendations for MSI testing based on consensus decisions from the ESMO Translational Research and Precision Medicine Working Group. The recommended first action to assess MSI/dMMR, decided by a consensus with a strong agreement, is the use of immunohistochemistry for the mismatch repair proteins MLH1, MSH2, MSH6, and PMS2. The second method of MSI/dMMR testing, also with a consensus of strong agreement, is the use of PCR-based assessment of microsatellite alterations using five microsatellite markers (including at least BAT-25 and BAT-26). Based on a consensus with a very strong agreement, the ESMO notes that NGS coupling MSI and TMB analysis may represent “a decisive tool for selecting patients for immunotherapy, for common or rare cancers not belonging to the spectrum of Lynch syndrome.” They note that the relationships between MSI, TMB, and PD-1/PD-L1 expression are both complex and different based on the type of tumor being assessed. They also note that the most important cancer types where MSI testing should be carried out using IHC to assess MSI-PCR or NGS are endometrial, intestinal (colorectal and small bowel), esophageal (adenocarcinomas and not squamous cell carcinoma), gastric, glioblastoma, and ovarian cancers (Luchini et al., 2019).
The ESMO has published clinical recommendations for the use of tumor multigene NGS in non-small cell lung cancer, cholangiocarcinoma, prostate, and ovarian cancers. They recommend to “test TMB in well- and moderately-differentiated neuroendocrine tumours (NETs), cervical, salivary, thyroid and vulvar cancers. . . as TMB-high predicted response to pembrolizumab in these cancers.” They advise that a large panel of genes could be ordered but that the benefit for the patient and the cost for the public health care system should be taken into consideration. Patient-specific recommendations are summarized in the table below (Mosele et al., 2020).
| Tumour Types |
General recommendations for daily practice |
| Lung adenocarcinoma |
Tumour multigene NGS to assess level I alterations. Larger panels can be used only on the basis of specific agreements with payers taking into account the overall cost of the strategy (drug includeda) and if they report accurate ranking of alterations. NGS can either be done on RNA or DNA, if it includes level I fusions in the panel. |
| Squamous cell lung cancers |
No current indication for tumour multigene NGS |
| Breast cancers |
No current indication for tumour multigene NGS |
| Colon cancers |
Multigene tumour NGS can be an alternative option to PCR if it does not result in additional cost. |
| Prostate cancers |
Multigene tumour NGS to assess level I alterations. Larger panels can be used only on the basis of specific agreements with payers taking into account the overall cost of the strategy and if they report accurate ranking of alterations. |
| Gastric cancers |
No current indication for tumour multigene NGS |
| Pancreatic cancers |
No current indication for tumour multigene NGS |
| Hepatocellular carcinoma |
No current indication for tumour multigene NGS |
| Cholangiocarcinoma |
Multigene tumour NGS could be recommended to assess level I alterations. Larger panels can be used only on the basis of specific agreements with payers taking into account the overall cost of the strategy (drug includeda) and if they report accurate ranking of alterations. RNA-based NGS can be used. |
| Others |
Tumour multigene NGS can be used in ovarian cancers to determine somatic BRCA1/2 mutations. In this latter case, larger panels can be used only on the basis of specific agreements with payers taking into account the overall cost of the strategy (drug includeda) and if they report accurate ranking of alterations. Large panel NGS can be used in carcinoma of unknown primary. It is recommended to determine TMB in cervical cancer, salivary cancer, thyroid cancers, well-to-moderately differentiated neuroendocrine tumours, vulvar cancer, pending drug access (and in TMB-high endometrial and SCL cancers if anti-PD1 antibody is not available otherwise). |
anti-PD1, anti-programmed cell death 1; DRUP, drug rediscovery protocol; ESMO, European Society for Medical Oncology; NGS, next-generation sequencing; SCL, small-cell lung cancer; TMB, tumour mutational burden.
a ESMO recommends using off-label drugs matched to genomics only if an access programme and a procedure of decision have been developed at the national or regional level, as illustrated by the DRUP programme.(Mosele et al., 2020)
College of American Pathologists (CAP)
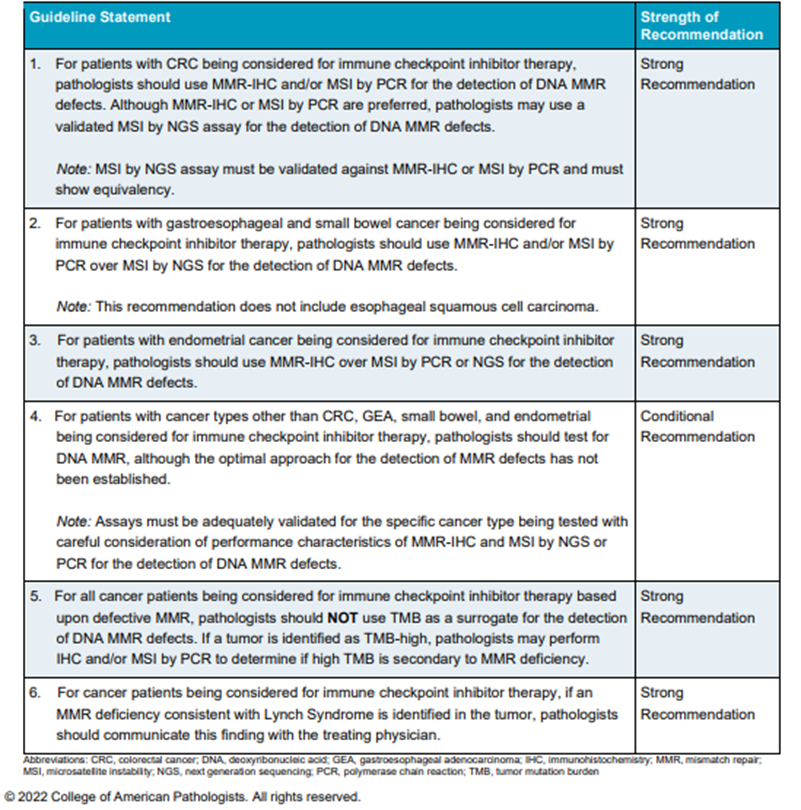
In 2022, the College of American Pathologists issued an evidence-based clinical guideline on the use of mismatch repair and microsatellite instability testing for immune checkpoint inhibitor therapy. These recommendations include guidance for patients with Lynch syndrome, as well as for patients with colorectal, endometrial, gastroesophageal, small bowel, and other types of cancers. Their summary of recommendations is captured below (CAP, 2022).
References
- Aggarwal, C., Ben-Shachar, R., Gao, Y., Hyun, S. W., Rivers, Z., Epstein, C., Kaneva, K., Sangli, C., Nimeiri, H., & Patel, J. (2023). Assessment of Tumor Mutational Burden and Outcomes in Patients With Diverse Advanced Cancers Treated With Immunotherapy. JAMA Netw Open, 6(5), e2311181. https://doi.org/10.1001/jamanetworkopen.2023.11181
- Alborelli, I., Leonards, K., Rothschild, S. I., Leuenberger, L. P., Savic Prince, S., Mertz, K. D., Poechtrager, S., Buess, M., Zippelius, A., Laubli, H., Haegele, J., Tolnay, M., Bubendorf, L., Quagliata, L., & Jermann, P. (2020). Tumor mutational burden assessed by targeted NGS predicts clinical benefit from immune checkpoint inhibitors in non-small cell lung cancer. J Pathol, 250(1), 19-29. https://doi.org/10.1002/path.5344
- Andre, T., Shiu, K. K., Kim, T. W., Jensen, B. V., Jensen, L. H., Punt, C., Smith, D., Garcia-Carbonero, R., Benavides, M., Gibbs, P., de la Fouchardiere, C., Rivera, F., Elez, E., Bendell, J., Le, D. T., Yoshino, T., Van Cutsem, E., Yang, P., Farooqui, M. Z. H., . . . Investigators, K.-. (2020). Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N Engl J Med, 383(23), 2207-2218. https://doi.org/10.1056/NEJMoa2017699
- Boland, C. R., Thibodeau, S. N., Hamilton, S. R., Sidransky, D., Eshleman, J. R., Burt, R. W., Meltzer, S. J., Rodriguez-Bigas, M. A., Fodde, R., Ranzani, G. N., & Srivastava, S. (1998). A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res, 58(22), 5248-5257. https://www.ncbi.nlm.nih.gov/pubmed/9823339
- Bonneville, R., Krook, M. A., Kautto, E. A., Miya, J., Wing, M. R., Chen, H. Z., Reeser, J. W., Yu, L., & Roychowdhury, S. (2017). Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis Oncol, 2017. https://doi.org/10.1200/PO.17.00073
- CAP. (2022). Mismatch Repair and Microsatellite Instability Testing for Immune Checkpoint Inhibitor Therapy. College of American Pathologists. https://www.cap.org/protocols-and-guidelines/cap-guidelines/current-cap-guidelines/mismatch-repair-and-microsatellite-instability-testing-for-immune-checkpoint-inhibitor-therapy
- Caris Life Sciences. (2024). MI Tumor Seek™. https://www.carislifesciences.com/products-and-services/molecular-profiling/
- Chalmers, Z. R., Connelly, C. F., Fabrizio, D., Gay, L., Ali, S. M., Ennis, R., Schrock, A., Campbell, B., Shlien, A., Chmielecki, J., Huang, F., He, Y., Sun, J., Tabori, U., Kennedy, M., Lieber, D. S., Roels, S., White, J., Otto, G. A., . . . Frampton, G. M. (2017). Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med, 9(1), 34. https://doi.org/10.1186/s13073-017-0424-2
- Cohen, R., Bennouna, J., Meurisse, A., Tournigand, C., De La Fouchardiere, C., Tougeron, D., Borg, C., Mazard, T., Chibaudel, B., Garcia-Larnicol, M. L., Svrcek, M., Vernerey, D., Menu, Y., & Andre, T. (2020). RECIST and iRECIST criteria for the evaluation of nivolumab plus ipilimumab in patients with microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: the GERCOR NIPICOL phase II study. J Immunother Cancer, 8(2). https://doi.org/10.1136/jitc-2020-001499
- FDA. (2017a). EVALUATION OF AUTOMATIC CLASS III DESIGNATION FOR MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets). https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN170058.pdf
- FDA. (2017b, November 15, 2017). FDA unveils a streamlined path for the authorization of tumor profiling tests alongside its latest product action https://www.fda.gov/news-events/press-announcements/fda-unveils-streamlined-path-authorization-tumor-profiling-tests-alongside-its-latest-product-action
- FDA. (2017c). FoundationOne CDx https://www.accessdata.fda.gov/cdrh_docs/pdf17/P170019B.pdf
- FDA. (2020a). FDA approves pembrolizumab for adults and children with TMB-H solid tumors. https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-adults-and-children-tmb-h-solid-tumors
- FDA. (2020b). FoundationOne Liquid CDx (F1 Liquid CDx). https://www.accessdata.fda.gov/cdrh_docs/pdf19/P190032B.pdf
- FDA. (2021, May 21, 2021). SUMMARY OF SAFETY AND EFFECTIVENESS DATA (SSED) Retrieved October 28, 2021 from https://www.accessdata.fda.gov/cdrh_docs/pdf20/P200010S001B.pdf
- Garofalo, A., Sholl, L., Reardon, B., Taylor-Weiner, A., Amin-Mansour, A., Miao, D., Liu, D., Oliver, N., MacConaill, L., Ducar, M., Rojas-Rudilla, V., Giannakis, M., Ghazani, A., Gray, S., Janne, P., Garber, J., Joffe, S., Lindeman, N., Wagle, N., . . . Van Allen, E. M. (2016). The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med, 8(1), 79. https://doi.org/10.1186/s13073-016-0333-9
- Georgiadis, A., Durham, J. N., Keefer, L. A., Bartlett, B. R., Zielonka, M., Murphy, D., White, J. R., Lu, S., Verner, E. L., Ruan, F., Riley, D., Anders, R. A., Gedvilaite, E., Angiuoli, S., Jones, S., Velculescu, V. E., Le, D. T., Diaz, L. A., & Sausen, M. (2019). Noninvasive Detection of Microsatellite Instability and High Tumor Mutation Burden in Cancer Patients Treated with PD-1 Blockade. Clinical Cancer Research. https://doi.org/10.1158/1078-0432.CCR-19-1372
- Guardant. (2024a). Blood-First, TissueNext Streamlined Biomarker Testing. Retrieved November 1, 2021 from https://guardant360cdx.com/tissuenext/
- Guardant. (2024b). TREATMENT SELECTION AND RESPONSE MONITORING FOR PATIENTS WITH ADVANCED CANCER. Retrieved October 28, 2021 from https://guardanthealth.com/solutions/#lunar-2
- Hellmann, M. D., Callahan, M. K., Awad, M. M., Calvo, E., Ascierto, P. A., Atmaca, A., Rizvi, N. A., Hirsch, F. R., Selvaggi, G., Szustakowski, J. D., Sasson, A., Golhar, R., Vitazka, P., Chang, H., Geese, W. J., & Antonia, S. J. (2018). Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell, 33(5), 853-861 e854. https://doi.org/10.1016/j.ccell.2018.04.001
- Hellmann, M. D., Ciuleanu, T. E., Pluzanski, A., Lee, J. S., Otterson, G. A., Audigier-Valette, C., Minenza, E., Linardou, H., Burgers, S., Salman, P., Borghaei, H., Ramalingam, S. S., Brahmer, J., Reck, M., O'Byrne, K. J., Geese, W. J., Green, G., Chang, H., Szustakowski, J., . . . Paz-Ares, L. (2018). Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med, 378(22), 2093-2104. https://doi.org/10.1056/NEJMoa1801946
- Hellmann, M. D., Nathanson, T., Rizvi, H., Creelan, B. C., Sanchez-Vega, F., Ahuja, A., Ni, A., Novik, J. B., Mangarin, L. M. B., Abu-Akeel, M., Liu, C., Sauter, J. L., Rekhtman, N., Chang, E., Callahan, M. K., Chaft, J. E., Voss, M. H., Tenet, M., Li, X. M., . . . Wolchok, J. D. (2018). Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell, 33(5), 843-852 e844. https://doi.org/10.1016/j.ccell.2018.03.018
- Illumina. (2024). TruSight Oncology 500. Retrieved October 28, 2021 from https://www.illumina.com/products/by-type/clinical-research-products/trusight-oncology-500.html?scid=2018-271doco175#productLongDescription
- Klempner, S. J., Fabrizio, D., Bane, S., Reinhart, M., Peoples, T., Ali, S. M., Sokol, E. S., Frampton, G., Schrock, A. B., Anhorn, R., & Reddy, P. (2020). Tumor Mutational Burden as a Predictive Biomarker for Response to Immune Checkpoint Inhibitors: A Review of Current Evidence. Oncologist, 25(1), e147-e159. https://doi.org/10.1634/theoncologist.2019-0244
- Le, D. T., Uram, J. N., Wang, H., Bartlett, B. R., Kemberling, H., Eyring, A. D., Skora, A. D., Luber, B. S., Azad, N. S., Laheru, D., Biedrzycki, B., Donehower, R. C., Zaheer, A., Fisher, G. A., Crocenzi, T. S., Lee, J. J., Duffy, S. M., Goldberg, R. M., de la Chapelle, A., . . . Diaz, L. A., Jr. (2015). PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med, 372(26), 2509-2520. https://doi.org/10.1056/NEJMoa1500596
- Luchini, C., Bibeau, F., Ligtenberg, M. J. L., Singh, N., Nottegar, A., Bosse, T., Miller, R., Riaz, N., Douillard, J. Y., Andre, F., & Scarpa, A. (2019). ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: a systematic review-based approach. Annals of Oncology, 30(8), 1232-1243. https://doi.org/10.1093/annonc/mdz116
- Marabelle, A., Fakih, M., Lopez, J., Shah, M., Shapira-Frommer, R., Nakagawa, K., Chung, H. C., Kindler, H. L., Lopez-Martin, J. A., Miller, W. H., Jr., Italiano, A., Kao, S., Piha-Paul, S. A., Delord, J. P., McWilliams, R. R., Fabrizio, D. A., Aurora-Garg, D., Xu, L., Jin, F., . . . Bang, Y. J. (2020). Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol, 21(10), 1353-1365. https://doi.org/10.1016/S1470-2045(20)30445-9
- Marabelle, A., Le, D. T., Ascierto, P. A., Di Giacomo, A. M., De Jesus-Acosta, A., Delord, J. P., Geva, R., Gottfried, M., Penel, N., Hansen, A. R., Piha-Paul, S. A., Doi, T., Gao, B., Chung, H. C., Lopez-Martin, J., Bang, Y. J., Frommer, R. S., Shah, M., Ghori, R., . . . Diaz, L. A., Jr. (2020). Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J Clin Oncol, 38(1), 1-10. https://doi.org/10.1200/JCO.19.02105
- Merino, D. M., McShane, L. M., Fabrizio, D., Funari, V., Chen, S. J., White, J. R., Wenz, P., Baden, J., Barrett, J. C., Chaudhary, R., Chen, L., Chen, W. S., Cheng, J. H., Cyanam, D., Dickey, J. S., Gupta, V., Hellmann, M., Helman, E., Li, Y., . . . Allen, J. (2020). Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer, 8(1). https://doi.org/10.1136/jitc-2019-000147
- Mosele, F., Remon, J., Mateo, J., Westphalen, C. B., Barlesi, F., Lolkema, M. P., Normanno, N., Scarpa, A., Robson, M., Meric-Bernstam, F., Wagle, N., Stenzinger, A., Bonastre, J., Bayle, A., Michiels, S., Bieche, I., Rouleau, E., Jezdic, S., Douillard, J. Y., . . . Andre, F. (2020). Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol, 31(11), 1491-1505. https://doi.org/10.1016/j.annonc.2020.07.014
- MSKCC. (2024). MSK-IMPACT: A Targeted Test for Mutations in Both Rare and Common Cancers. Retrieved October 20, 2021 from https://www.mskcc.org/msk-impact
- NCCN. (2024a, August 20, 2024). NCCN Clinical Practice Guidelines in Ampullary Adenocarcinoma Version 1.2025 — August 20, 2024. https://www.nccn.org/professionals/physician_gls/pdf/ampullary.pdf
- NCCN. (2024b, April 30, 2024). NCCN Clinical Practice Guidelines in Oncology - Pancreatic Adenocarcinoma Version 2.2024 https://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf
- NCCN. (2024c). NCCN Clinical Practice Guidelines in Oncology - Penile Cancer Version 1.2025 — November 1, 2024. Retrieved November 05, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/penile.pdf
- NCCN. (2024d, September 24, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Biliary Tract Cancers Version 3.2024 — September 24, 2024. https://www.nccn.org/professionals/physician_gls/pdf/btc.pdf
- NCCN. (2024e, August 20, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Bone Cancer Version 1.2025 — August 20, 2024. Retrieved November 05, 2022 from https://www.nccn.org/professionals/physician_gls/PDF/bone.pdf
- NCCN. (2024f, October 15, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Breast Cancer Version 5.2024 —October 15, 2024. Retrieved November 5, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf
- NCCN. (2024g, September 24, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Cervical Cancer Version 3.2024 Retrieved October 12, 2021 from https://www.nccn.org/professionals/physician_gls/pdf/cervical.pdf
- NCCN. (2024h, August 22, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Colon Cancer Version 5.2024 Retrieved November 5, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf
- NCCN. (2024i, July 30, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Esophageal and Esophagogastric Junction Cancers Version Version 4.2024 Retrieved November 5, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/esophageal.pdf
- NCCN. (2024j, August 12, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Gastric Cancer Version 4.2024 Retrieved November 5, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/gastric.pdf
- NCCN. (2024k, August 2, 2023). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Neuroendocrine and Adrenal Tumors Version 2.2024 — August 1, 2024. Retrieved November 05, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/neuroendocrine.pdf
- NCCN. (2024l, October 15, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Non-Small Cell Lung Cancer Version 11.2024. https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf
- NCCN. (2024m, September 11, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Occult Primary (Cancer of Unknown Primary [CUP]) Version 2.2025 — September 11, 2024. Retrieved November 5, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/occult.pdf
- NCCN. (2024n, July 15, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Ovarian Cancer, Including Fallopian Tube Cancer and Primary Peritoneal Cancer Version 3.2024. Retrieved November 05, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/ovarian.pdf
- NCCN. (2024o, May 17, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Prostate Cancer Version 4.2024. Retrieved November 05, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
- NCCN. (2024p, August 22, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Rectal Cancer Version 4.2024 Retrieved November 05, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/rectal.pdf
- NCCN. (2024q, September 13, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Small Bowel Adenocarcinoma Version 5.2024 Retrieved November 05, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/small_bowel.pdf
- NCCN. (2024r, October 29, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Testicular Cancer Version 2.2024 — October 29, 2024. Retrieved November 05, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/testicular.pdf
- NCCN. (2024s, August 19, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Thyroid Carcinoma Version 4.2024 — August 19, 2024. Retrieved November 5, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/thyroid.pdf
- NCCN. (2024t, September 20, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Uterine Neoplasms Version 3.2024 — September 20, 2024. Retrieved October 12, 2021 from https://www.nccn.org/professionals/physician_gls/pdf/uterine.pdf
- NCCN. (2024u, May 1, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines): Vulvar Cancer Version 4.2024 — May 1, 2024. Retrieved November 05, 2022 from https://www.nccn.org/professionals/physician_gls/pdf/vulvar.pdf
- NCCN. (2024v, March 4, 2024). NCCN Clinical Practice Guidelines in Oncology Anal Carcinoma NCCN Evidence Blocks TM Version 1.2024 — March 4, 2024. https://www.nccn.org/professionals/physician_gls/pdf/anal_blocks.pdf
- NCCN. (2024w, October 24, 2024). NCCN Clinical Practice Guidelines in Oncology Head and Neck Cancers Version 5.2024. https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf
- NCCN. (2024x, September 24, 2024). NCCN Clinical Practice Guidelines in Oncology in Hepatocellular Carcinoma Version 3.2024 — September 24, 2024. https://www.nccn.org/professionals/physician_gls/pdf/hcc.pdf
- NCCN. (2024y, February 26, 2024). NCCN Clinical Practice Guidelines in Oncology in Pediatric Central Nervous System Cancers Version 1.2024 — February 26, 2024. https://www.nccn.org/professionals/physician_gls/pdf/ped_cns.pdf
- NCCN. (2024z, September 27, 2024). NCCN Clinical Practice Guidelines in Oncology in Soft Tissue Sarcoma Version 3.2024. https://www.nccn.org/professionals/physician_gls/pdf/sarcoma.pdf
- NCI. (n.d.). MSI-H cancer. https://www.cancer.gov/publications/dictionaries/cancer-terms/def/msi-h-cancer
- Nojadeh, J. N., Behrouz Sharif, S., & Sakhinia, E. (2018). Microsatellite instability in colorectal cancer. EXCLI J, 17, 159-168. https://doi.org/10.17179/excli2017-948
- Overman, M. J., McDermott, R., Leach, J. L., Lonardi, S., Lenz, H. J., Morse, M. A., Desai, J., Hill, A., Axelson, M., Moss, R. A., Goldberg, M. V., Cao, Z. A., Ledeine, J. M., Maglinte, G. A., Kopetz, S., & Andre, T. (2017). Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol, 18(9), 1182-1191. https://doi.org/10.1016/S1470-2045(17)30422-9
- Qiagen. (2024). QIAseq Tumor Mutational Burden Panels. Retrieved October 28, 2021 from https://www.qiagen.com/us/products/next-generation-sequencing/dna-sequencing/targeted-dna-panels/qiaseq-tumor-mutational-burden-panels/
- Quy, P. N., Kanai, M., Fukuyama, K., Kou, T., Kondo, T., Yamamoto, Y., Matsubara, J., Hiroshima, A., Mochizuki, H., Sakuma, T., Kamada, M., Nakatsui, M., Eso, Y., Seno, H., Masui, T., Takaori, K., Minamiguchi, S., Matsumoto, S., & Muto, M. (2019). Association Between Preanalytical Factors and Tumor Mutational Burden Estimated by Next-Generation Sequencing-Based Multiplex Gene Panel Assay. Oncologist, 24(12), e1401-e1408. https://doi.org/10.1634/theoncologist.2018-0587
- Ready, N., Hellmann, M. D., Awad, M. M., Otterson, G. A., Gutierrez, M., Gainor, J. F., Borghaei, H., Jolivet, J., Horn, L., Mates, M., Brahmer, J., Rabinowitz, I., Reddy, P. S., Chesney, J., Orcutt, J., Spigel, D. R., Reck, M., O'Byrne, K. J., Paz-Ares, L., . . . Ramalingam, S. S. (2019). First-Line Nivolumab Plus Ipilimumab in Advanced Non-Small-Cell Lung Cancer (CheckMate 568): Outcomes by Programmed Death Ligand 1 and Tumor Mutational Burden as Biomarkers. J Clin Oncol, 37(12), 992-1000. https://doi.org/10.1200/JCO.18.01042
- Ritterhouse, L. L. (2019). Tumor mutational burden. Cancer Cytopathol, 127(12), 735-736. https://doi.org/10.1002/cncy.22174
- Rizvi, N. A., Hellmann, M. D., Snyder, A., Kvistborg, P., Makarov, V., Havel, J. J., Lee, W., Yuan, J., Wong, P., Ho, T. S., Miller, M. L., Rekhtman, N., Moreira, A. L., Ibrahim, F., Bruggeman, C., Gasmi, B., Zappasodi, R., Maeda, Y., Sander, C., . . . Chan, T. A. (2015). Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science, 348(6230), 124-128. https://doi.org/10.1126/science.aaa1348
- Samstein, R. M., Lee, C. H., Shoushtari, A. N., Hellmann, M. D., Shen, R., Janjigian, Y. Y., Barron, D. A., Zehir, A., Jordan, E. J., Omuro, A., Kaley, T. J., Kendall, S. M., Motzer, R. J., Hakimi, A. A., Voss, M. H., Russo, P., Rosenberg, J., Iyer, G., Bochner, B. H., . . . Morris, L. G. T. (2019). Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet, 51(2), 202-206. https://doi.org/10.1038/s41588-018-0312-8
- Si, H., Kuziora, M., Quinn, K. J., Helman, E., Ye, J., Liu, F., Scheuring, U., Peters, S., Rizvi, N. A., Brohawn, P. Z., Ranade, K., Higgs, B. W., Banks, K. C., Chand, V. K., & Raja, R. (2021). A Blood-based Assay for Assessment of Tumor Mutational Burden in First-line Metastatic NSCLC Treatment: Results from the MYSTIC Study. Clin Cancer Res, 27(6), 1631-1640. https://doi.org/10.1158/1078-0432.CCR-20-3771
- Singal, G., Miller, P. G., Agarwala, V., Li, G., Kaushik, G., Backenroth, D., Gossai, A., Frampton, G. M., Torres, A. Z., Lehnert, E. M., Bourque, D., O'Connell, C., Bowser, B., Caron, T., Baydur, E., Seidl-Rathkopf, K., Ivanov, I., Alpha-Cobb, G., Guria, A., . . . Miller, V. A. (2019). Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA, 321(14), 1391-1399. https://doi.org/10.1001/jama.2019.3241
- Snyder, A., Makarov, V., Merghoub, T., Yuan, J., Zaretsky, J. M., Desrichard, A., Walsh, L. A., Postow, M. A., Wong, P., Ho, T. S., Hollmann, T. J., Bruggeman, C., Kannan, K., Li, Y., Elipenahli, C., Liu, C., Harbison, C. T., Wang, L., Ribas, A., . . . Chan, T. A. (2014). Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med, 371(23), 2189-2199. https://doi.org/10.1056/NEJMoa1406498
- Stahler, A., Stintzing, S., von Einem, J. C., Westphalen, C. B., Heinrich, K., Kramer, N., Michl, M., Modest, D. P., von Weikersthal, L. F., Decker, T., Kiani, A., Heintges, T., Kahl, C., Kullmann, F., Scheithauer, W., Moehler, M., Kaiser, F., Kirchner, T., Jung, A., & Heinemann, V. (2020). Single-nucleotide variants, tumour mutational burden and microsatellite instability in patients with metastatic colorectal cancer: Next-generation sequencing results of the FIRE-3 trial. Eur J Cancer, 137, 250-259. https://doi.org/10.1016/j.ejca.2020.07.003
- Van Allen, E. M., Miao, D., Schilling, B., Shukla, S. A., Blank, C., Zimmer, L., Sucker, A., Hillen, U., Foppen, M. H. G., Goldinger, S. M., Utikal, J., Hassel, J. C., Weide, B., Kaehler, K. C., Loquai, C., Mohr, P., Gutzmer, R., Dummer, R., Gabriel, S., . . . Garraway, L. A. (2015). Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science, 350(6257), 207-211. https://doi.org/10.1126/science.aad0095
- Wei, B., Kang, J., Kibukawa, M., Arreaza, G., Maguire, M., Chen, L., Qiu, P., Lang, L., Aurora-Garg, D., Cristescu, R., & Levitan, D. (2022). Evaluation of the TruSight Oncology 500 Assay for Routine Clinical Testing of Tumor Mutational Burden and Clinical Utility for Predicting Response to Pembrolizumab. J Mol Diagn. https://doi.org/10.1016/j.jmoldx.2022.01.008
- Willis, C., Fiander, M., Tran, D., Korytowsky, B., Thomas, J. M., Calderon, F., Zyczynski, T. M., Brixner, D., & Stenehjem, D. D. (2019). Tumor mutational burden in lung cancer: a systematic literature review. Oncotarget, 10(61), 6604-6622. https://doi.org/10.18632/oncotarget.27287
- Willis, J., Lefterova, M. I., Artyomenko, A., Kasi, P. M., Nakamura, Y., Mody, K., Catenacci, D. V. T., Fakih, M., Barbacioru, C., Zhao, J., Sikora, M., Fairclough, S. R., Lee, H., Kim, K.-M., Kim, S. T., Kim, J., Gavino, D., Benavides, M., Peled, N., . . . Odegaard, J. I. (2019). Validation of Microsatellite Instability Detection Using a Comprehensive Plasma-Based Genotyping Panel. Clinical Cancer Research. https://doi.org/10.1158/1078-0432.CCR-19-1324
- Woodhouse, R., Li, M., Hughes, J., Delfosse, D., Skoletsky, J., Ma, P., Meng, W., Dewal, N., Milbury, C., Clark, T., Donahue, A., Stover, D., Kennedy, M., Dacpano-Komansky, J., Burns, C., Vietz, C., Alexander, B., Hegde, P., & Dennis, L. (2020). Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One, 15(9), e0237802. https://doi.org/10.1371/journal.pone.0237802
- Wu, H. X., Wang, Z. X., Zhao, Q., Wang, F., & Xu, R. H. (2019). Designing gene panels for tumor mutational burden estimation: the need to shift from 'correlation' to 'accuracy'. J Immunother Cancer, 7(1), 206. https://doi.org/10.1186/s40425-019-0681-2
- Yarchoan, M., Albacker, L. A., Hopkins, A. C., Montesion, M., Murugesan, K., Vithayathil, T. T., Zaidi, N., Azad, N. S., Laheru, D. A., Frampton, G. M., & Jaffee, E. M. (2019). PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight, 4(6). https://doi.org/10.1172/jci.insight.126908
- Yarchoan, M., Hopkins, A., & Jaffee, E. M. (2017). Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med, 377(25), 2500-2501. https://doi.org/10.1056/NEJMc1713444
- Zhao, P., Li, L., Jiang, X., & Li, Q. (2019). Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol, 12(1), 54. https://doi.org/10.1186/s13045-019-0738-1
Coding Section
| Code | Number | Description |
| CPT | 81301 | Microsatellite instability analysis (e.g., hereditary non-polyposis colorectal cancer, Lynch syndrome) of markers for mismatch repair deficiency (e.g., BAT25, BAT26), includes comparison of neoplastic and normal tissue, if performed |
| 81457 | Solid organ neoplasm, genomic sequence analysis panel, interrogation for sequence variants; DNA analysis, microsatellite instability | |
| 81458 | Solid organ neoplasm, genomic sequence analysis panel, interrogation for sequence variants; DNA analysis, copy number variants and microsatellite instability | |
| 81459 | Solid organ neoplasm, genomic sequence analysis panel, interrogation for sequence variants; DNA analysis or combined DNA and RNA analysis, copy number variants, microsatellite instability, tumor mutation burden, and rearrangements | |
| 81463 (effective 01/01/2024) |
Solid organ neoplasm, genomic sequence analysis panel, cell-free nucleic acid (e.g., plasma), interrogation for sequence variants; DNA analysis, copy number variants, and microsatellite instability |
|
| 81464 (effective 01/01/2024) | Solid organ neoplasm, genomic sequence analysis panel, cell-free nucleic acid (e.g., plasma), interrogation for sequence variants; DNA analysis or combined DNA and RNA analysis, copy number variants, microsatellite instability, tumor mutation burden, and rearrangements |
|
| 81479 |
Unlisted molecular pathology procedure |
|
| 0037U |
Targeted genomic sequence analysis, solid organ neoplasm, DNA analysis of 324 genes, interrogation for sequence variants, gene copy number amplifications, gene rearrangements, microsatellite instability and tumor mutational burden |
|
| 0048U |
Oncology (solid organ neoplasia), DNA, targeted sequencing of protein-coding exons of 468 cancer-associated genes, including interrogation for somatic mutations and microsatellite instability, matched with normal specimens, utilizing formalin-fixed paraffin-embedded tumor tissue, report of clinically significant mutation(s) |
|
| 0211U |
Oncology (pan-tumor), DNA and RNA by next-generation sequencing, utilizing formalin-fixed paraffin-embedded tissue, interpretative report for single nucleotide variants, copy number alterations, tumor mutational burden, and microsatellite instability, with therapy association |
|
| 0239U |
Targeted genomic sequence analysis panel, solid organ neoplasm, cell-free DNA, analysis of 311 or more genes, interrogation for sequence variants, including substitutions, insertions, deletions, select rearrangements, and copy number variations |
|
| 0242U |
Targeted genomic sequence analysis panel, solid organ neoplasm, cell-free circulating DNA analysis of 55 – 74 genes, interrogation for sequence variants, gene copy number amplifications, and gene rearrangements |
|
| 0244U |
Oncology (solid organ), DNA, comprehensive genomic profiling, 257 genes, interrogation for single-nucleotide variants, insertions/deletions, copy number alterations, gene rearrangements, tumor-mutational burden and microsatellite instability, utilizing formalin-fixed paraffin-embedded tumor tissue |
|
| 0250U |
Oncology (solid organ neoplasm), targeted genomic sequence DNA analysis of 505 genes, interrogation for somatic alterations (SNVs [single nucleotide variant], small insertions and deletions, one amplification, and four translocations), microsatellite instability and tumor-mutation burden |
|
| 0326U |
Targeted genomic sequence analysis panel, solid organ neoplasm, cell-free circulating DNA analysis of 83 or more genes, interrogation for sequence variants, gene copy number amplifications, gene rearrangements, microsatellite instability and tumor mutational |
|
| 0329U | Oncology (neoplasia), exome and transcriptome sequence analysis for sequence variants, gene copy number amplifications and deletions, gene rearrangements, microsatellite instability and tumor mutational burden utilizing DNA and RNA from tumor with DNA from normal blood or saliva for subtraction, report of clinically significant mutation(s) with therapy associations |
|
| 0334U | Oncology (solid organ), targeted genomic sequence analysis, formalin-fixed paraffin-embedded (FFPE) tumor tissue, DNA analysis, 84 or more genes, interrogation for sequence variants, gene copy number amplifications, gene rearrangements, microsatellite instability and tumor mutational burden Protietary test: Guardant360 TissueNext™ Lab/Manufacturer: Guardant Health Inc. |
|
| 0379U | Targeted genomic sequence analysis panel, solid organ neoplasm, DNA (523 genes) and RNA (55 genes) by next-generation sequencing, interrogation for sequence variants, gene copy number amplifications, gene rearrangements, microsatellite instability, and tumor mutational burden Proprietary test: Solid Tumor Expanded Panel Lab/Manufacturer: Quest Diagnostics® |
|
| 0391U (effective 07/01/2023) | Oncology (solid tumor), DNA and RNA by next-generation sequencing, utilizing formalin-fixed paraffin-embedded (FFPE) tissue, 437 genes, interpretive report for single nucleotide variants, splicesite variants, insertions/deletions, copy number alterations, gene fusions, tumor mutational burden, and microsatellite instability, with algorithm quantifying immunotherapy response score Proprietary test: Strata Select™ Lab/Manufacturer: Strata Oncology, Inc. |
|
| 0409U (effective 10/01/2023) | Oncology (solid tumor), DNA (80 genes) and RNA (36 genes), by next-generation sequencing from plasma, including single nucleotide variants, insertions/deletions, copy number alterations, microsatellite instability, and fusions, report showing identified mutations with clinical actionability Proprietary test: LiquidHALLMARK ® Lab/Manufacturer: Lucene Health, Inc. |
|
| 0473U (effective date 07/01/2024) | Oncology (solid tumor), next generation sequencing (NGS) of DNA from formalin-fixed paraffin embedded (FFPE) tissue with comparative sequence analysis from a matched normal specimen (blood or saliva), 648 genes, interrogation for sequence variants, insertion and deletion alterations, copy number variants, rearrangements, microsatellite instability, and tumor-mutation burden Protietary test: xT CDx Lab/Manufacturer: Tempus AI Inc. |
|
| 0487U (effective date 10/01/2024) | Oncology (solid tumor), cell-free circulating DNA, targeted genomic sequence analysis panel of 84 genes, interrogation for sequence variants, aneuploidycorrected gene copy number amplifications and losses, gene rearrangements, and microsatellite instability Protietary test: Northstar SelectTM Lab/Manufacturer: BillionToOne Laboratory |
|
| 0512U (effective date 10/01/2024) | Oncology (prostate), augmentative algorithmic analysis of digitized whole-slide imaging of histologic features for microsatellite instability (MSI) status, formalin-fixed paraffinembedded (FFPE) tissue, reported as increased or decreased probability of MSI-high (MSI-H) |
|
| 0513U (effective date 10/01/2024) | Oncology (prostate), augmentative algorithmic analysis of digitized whole-slide imaging of histologic features for microsatellite instability (MSI) status, and homologous recombination deficiency (HRD) status, formalinfixed paraffin-embedded (FFPE) tissue, reported as increased or decreased probability of each biomarker Protietary test: Tempus p-Prostate Lab/Manufacturer: Tempus AI, Inc |
|
| 0530U (effective date 01/01/2025) | Oncology (pan-solid tumor), ctDNA, utilizing plasma, nextgeneration sequencing (NGS) of 77 genes, 8 fusions, microsatellite instability, and tumor mutation burden, interpretative report for single nucleotide variants, copynumber alterations, with therapy association Protietary test: LiquidHALLMARK® Lab/Manufacturer: Lucence Health, Inc, Lucence Health, Inc |
|
| 0538U (effective date 04/01/2025) | Oncology (solid tumor), next?generation targeted sequencing analysis, formalin-fixed paraffin?embedded (FFPE) tumor tissue, DNA analysis of 600 genes, interrogation for single-nucleotide variants, insertions/deletions, gene rearrangements, and copy number alterations, microsatellite instability, tumor mutation burden, reported as actionable variant | |
| 0539U (effective date 04/01/2025) | Oncology (solid tumor), cellfree circulating tumor DNA (ctDNA), 152 genes, nextgeneration sequencing, interrogation for singlenucleotide variants, insertions/deletions, gene rearrangements, copy number alterations, and microsatellite instability, using whole-blood samples, mutations with clinical actionability reported as actionable variant |
|
| 0543U (effective date 04/01/2025) | Oncology (solid tumor), next?generation sequencing of DNA from formalin-fixed paraffin-embedded (FFPE) tissue of 517 genes, interrogation for single?nucleotide variants, multi?nucleotide variants, insertions and deletions from DNA, fusions in 24 genes and splice variants in 1 gene from RNA, and tumor mutation burden |
Procedure and diagnosis codes on Medical Policy documents are included only as a general reference tool for each policy. They may not be all-inclusive.
This medical policy was developed through consideration of peer-reviewed medical literature generally recognized by the relevant medical community, U.S. FDA approval status, nationally accepted standards of medical practice and accepted standards of medical practice in this community and other nonaffiliated technology evaluation centers, reference to federal regulations, other plan medical policies, and accredited national guidelines.
"Current Procedural Terminology © American Medical Association. All Rights Reserved"
History From 2021 Forward
| 02/17/2025 | Interim review, updating table of solid tumors, rationale, references, and coding. |
| 02/11/2025 | Updated coding section. Added codes 0538U, 0539U and 0543U. These codes will be effective 04/01/2025. No other changes. |
| 11/13/2024 | Updating CPT coding. Code 0428U will be deleted on 01/01/2025. Also adding code 0530U to be effective on 01/01/2025. |
| 09/05/2024 | Updated CPT coding. Added codes 0487U, 0512U and 0513U (effective 10/01/2024). No change in policy intent. |
| 08/13/2024 | Annual review updating TMB/MSI targeted therapy indications table. Also updating rationale and references and adding 0329U to coding. |
| 06/25/2024 | Interim review to add PLA code 0473U to coding section. |
| 05/01/2024 | Interim review to add NSCLC to table two. |
| 01/24/2024 | Annual review, updating policy / note 2 to include ampullary carcinoma and pediatric central nervous system cancer. Updating hepatobiliary cancer criteria to be reflected as biliary tract cancers. Also updating table of terminology, rationale and references. |
| 01/11/2024 | Added code 81463 & 81464 effective 01/01/2024. |
| 09/11/2023 | Updating coding section. Adding CPT code 0409U effective 10/01/2023. No other change made. |
| 07/17/2023 | Interim review updating policy coverage criteria for cervical and colon cancer based on NCCN updates. Also updating notes, rational, and references and adding PLA 0391U. |
| 06/05/2023 | Adding Code 0391U to coding section. No other changes made. |
| 01/25/2023 | Annual review, policy updated for clarity and consistency with coverage updated to mirror NCCN guidelines. also updating description, rationale, references, notes/guidelines and adding PLA code 0334U. |
| 08/11/2022 | Returning policy to July review month. Updating description, rationale, references and coding. Policy reformatted for clarity. |
| 06/15/2022 | Adding code 0326U |
| 01/13/2022 |
New Policy |