Minimal Residual Disease - CAM 251
Description
Minimal residual disease, also called measurable residual disease or MRD, refers to the subclinical levels of residual diseases, such as acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), and multiple myeloma (MM) (Horton & Steuber, 2023; Rajkumar, 2023; Stock & Estrov, 2022a, 2022b). MRD is a postdiagnosis, prognostic indicator that can be used for risk stratification and to guide therapeutic options when used alongside other clinical and molecular data (Schuurhuis et al., 2018). Many different techniques have been developed to detect residual disease. However, PCR-based techniques, multicolor flow cytometry, and deep sequencing-based MRD generally provide the best sensitivity, specificity, reproducibility, and applicability compared to other techniques such as fluorescence in situ hybridization (FISH), Southern blotting, or cell culture (Stock & Estrov, 2022b).
Regulatory Status
The FDA approved ClonoSEQ for marketing in 2018. The FDA notes that the test was approved through its “de novo premarket review pathway” and authorized Adaptive Biotechnologies to market this assay (FDA, 2018b). In its Decision Summary, the FDA states that the “The clonoSEQ Assay is an in vitro diagnostic that uses multiplex polymerase chain reaction (PCR) and next-generation sequencing (NGS) to identify and quantify rearranged IgH (VDJ), IgH (DJ), IgK, and IgL receptor gene sequences, as well as translocated BCL1/IgH (J) and BCL2/IgH (J) sequences in DNA extracted from bone marrow from patients with B-Cell acute lymphoblastic leukemia (ALL) or multiple myeloma (MM). The clonoSEQ Assay measures minimal residual disease (MRD) to monitor changes in burden of disease during and after treatment (FDA, 2018a)."
A search of the FDA device database on July 9, 2020, of “minimal residual disease” and “MRD” resulted in no additional pertinent results. Additionally, many labs have developed specific tests that they must validate and perform in-house. These laboratory-developed tests (LDTs) are regulated by the Centers for Medicare & Medicaid Services (CMS) as high-complexity tests under the Clinical Laboratory Improvement Amendments of 1988 (CLIA ’88). As an LDT, the U.S. Food and Drug Administration has not approved or cleared this test; however, FDA clearance or approval is not currently required for clinical use.
Policy
Application of coverage criteria is dependent upon an individual’s benefit coverage at the time of the request.
- For individuals with multiple myeloma (MM), chronic lymphocytic leukemia (CLL), or small lymphocytic lymphoma (SLL), minimal residual disease (MRD) testing by multiparameter flow cytometry or next-generation sequencing (NGS) is considered MEDICALLY NECESSARY.
- For individuals with acute myeloid leukemia (AML) or acute lymphoblastic leukemia (ALL), MRD testing by multiparameter flow cytometry, PCR-based techniques, or NGS is considered MEDICALLY NECESSARY.
- For individuals with a diagnosis of human papilloma virus (HPV)-positive oropharyngeal squamous cell carcinoma (OPSCC) who are being monitored for disease recurrence (at least three months after completion of initial definitive therapy), blood-based detection of tumor tissue modified viral (TTMV)-HPV DNA fragments (i.e., NavDx®) is considered MEDICALLY NECESSARY once every three months.
The following does not meet coverage criteria due to a lack of available published scientific literature confirming that the test(s) is/are required and beneficial for the diagnosis and treatment of an individual’s illness.
- For all situations not addressed above (e.g., MRD testing for all other solid tumors), MRD testing by multiparameter flow cytometry, PCR, or NGS is considered NOT MEDICALLY NECESSARY.
Table of Terminology
| Term |
Definition |
| ABL |
Abelson tyrosine kinase gene |
| AL |
Acute leukemia |
| ALL |
Acute lymphoblastic leukemia |
| ALL |
Pediatric acute lymphoblastic leukemia |
| AML |
Acute myeloid leukemia |
| AML1 |
Acute myeloid leukemia 1 protein gene |
| ASCO |
American Society of Clinical Oncology |
| ASCT |
Autologous Stem Cell Transplantation |
| ASH |
American Society of Hematology |
| B-ALL |
B-cell acute lymphoblastic leukemia |
| BCL1 |
B-cell lymphoma 1 gene |
| BCL2 |
B-cell lymphoma 2 gene |
| BCP |
B-cell precursor |
| BCR |
Break point cluster gene |
| BM |
Bone marrow |
| BMA |
Bone marrow aspirate |
| BMR |
Bone marrow relapse |
| BRAF |
Proto-oncogene B-Raf/v-Raf murine sarcoma viral oncogene homolog B |
| CA |
Cytogenetic abnormalities |
| CAP |
College of American Pathologists |
| CAR |
Post-chimeric antigen receptor |
| CBF |
Core binding factor |
| CBFB-MYH11 |
Cord binding factor subunit beta-myosin 11 fusion gene |
| CCO |
Cancer Care Ontario |
| CD19 |
Cluster of differentiation 19 |
| CD20 |
Cluster of differentiation 20 |
| CD43 |
Cluster of differentiation 43 |
| CD49b |
Cluster of differentiation 49b |
| CD5 |
Cluster of differentiation 5 |
| CD81 |
Cluster of differentiation 81 |
| 2-CldA |
Cladribine |
| CLIA |
Clinical Laboratory Improvement Amendments |
| CLL |
Chronic lymphocytic leukemia |
| CMS |
Centers for Medicare & Medicaid Services |
| CR |
Complete response/complete remission |
| CT | Computed Tomography |
| ctDNA |
Circulating tumor deoxyribonucleic acid |
| CV |
Coefficients of variance |
| DCF |
Pentostatin |
| DFCI |
Dana Farber Cancer Institute |
| DFS |
Disease Free Survival |
| DJ |
Diversity and joining segment rearrangement |
| DNA |
Deoxyribonucleic acid |
| E2A |
Transcription factor E2-alpha gene |
| EFS |
Event free survival |
| ELN |
European LeukemiaNet |
| EMN |
European Myeloma Network |
| EOCT |
End of combination therapy |
| ERIC |
European Research Initiative on chronic lymphocytic leukemia |
| ESMO |
European Society for Medical Oncology |
| EWS |
Ewing sarcoma |
| FC |
Flow cytometry |
| FCR |
Fludarabine cyclophosphamide and rituximab |
| FDA |
Food and Drug Administration |
| FISH |
Fluorescence in situ hybridization |
| FUM3 |
Follow up month 3 |
| gDNA |
Genomic deoxyribonucleic acid |
| HPV | Human papillomavirus |
| HR-NB |
High-risk neuroblastoma |
| HSCT |
Haematopoietic stem cell transplantation |
| HTS |
High-throughput sequencing |
| IgH |
Immunoglobulin heavy chain |
| IGH |
Immunoglobulin heavy chain gene |
| IgK |
Immunoglobulin K |
| IgL |
Immunoglobulin L |
| IHC |
Immunohistochemistry |
| IMWG |
International Myeloma Working Group |
| iwCLL |
International Workshop on Chronic Lymphocytic Leukemia |
| J |
Joining |
| KIT |
KIT proto-oncogene receptor tyrosine kinase |
| LDTs |
Laboratory developed tests |
| MFC/mFC |
Multicolor flow cytometry |
| MFC |
Mutliparameter flow cytometry |
| MM |
Multiple myeloma |
| mpFC |
Multi parametic flow cytometry |
| MRD |
Minimal residual disease |
| MRD |
Measurable residual disease |
| NCCN |
National Comprehensive Cancer Network |
| NDMM |
Newly diagnosed multiple myeloma |
| NGF |
Next-generation flow cytometry |
| NGS |
Next generation sequencing |
| NGS-MRD |
Next-generation sequencing-minimal residual disease |
| non-TBI |
Non-total body irradiation |
| NPV |
Negative Predictive Value |
| NPM1mut |
Nucleophosmin 1 gene mutant allele |
| OS |
Overall survival |
| OPSCC | Oropharyngeal squamous cell carcinoma |
| PBX1 |
Pre-B-cell leukemia transcription factor 1 gene |
| PCR |
Polymerase chain reaction |
| PET-CT |
Positron emission tomography -computed tomography |
| PFS |
Progression-free survival |
| PML-RARA |
Promyelocytic leukemia-retinoic acid receptor alpha fusion gene |
| PPV |
Positive predictive value |
| QALYs |
Quality adjusted life years |
| RQ-PCR/RT-qPCR |
Real-time quantitative polymerase chain reaction |
| RRMM |
Relapsed refractory multiple myeloma |
| RT-PCR |
Reverse transcriptase polymerase chain reaction |
| RUNX1-RUNX1T1 |
Runt-related transcription factor 1- RUNX1 partner transcriptional co-repressor 1 gene |
| sCR |
Stringent complete remission |
| SR |
Standard risk |
| T-ALL |
T-cell acute lymphocytic leukemia |
| TBI |
Total body irradiation |
| TCR |
T cell receptor |
| TEL |
Translocation-Ets-leukemia virus gene |
| TRG |
T cell receptor gamma locus gene |
| TTMV | Tumor Tissue modified viral |
| uMRD |
Undetectable minimal residual disease |
| VDJ |
Variable-diversity-joining |
| VGPR |
Very good partial response |
| WGS |
Whole genome sequencing |
Rationale
The goal of treating cancer has traditionally been “complete remission” (or response), defined as “absence of visible tumor” based on techniques, such as imaging and histological examination of tissue (Luskin et al., 2018). However, some cancer cells may remain undetected due to a lack of sensitivity of conventional methods, leading to relapse. This subclinical amount of cancer cells is referred to as minimal residual disease or MRD (Bai et al., 2018). While many techniques have been developed to determine MRD, multicolor flow cytometry and PCR-based, including next generation sequencing (NGS), MRD techniques are the most commonly used (Rai & Stilgenbauer, 2024; Stock & Estrov, 2022b).
Multicolor flow cytometry (MFC), also known as multiparameter flow cytometry, can be used to identify MRD by measuring for the aberrant expression of antigens on cancer cells. MFC uses lasers of different colors to simultaneously determine specific immunophenotypic features of the cells within a sample. “Classic flow cytometry techniques using four to six colors have limited sensitivity and specificity for MRD detection. Current flow cytometry techniques use six to eight colors to assess MRD with a sensitivity which is approximately 10-4, or about 0.5 to 1 log lower than that of polymerase chain reaction” (PCR) (Stock & Estrov, 2022b).
Polymerase chain reaction-based MRD techniques, including NGS, amplify sequences of DNA unique to the cancerous cell. These techniques have amazing sensitivity. In fact, real-time quantitative PCR can be used to detect a single cancerous cell from 104 – 105 cells (Brüggemann et al., 2006; Stock & Estrov, 2022b). The targets of amplification can include T cell receptor (TCR) gene rearrangements, the immunoglobulin heavy chain (IgH), or even fusion-gene transcripts (Del Giudice et al., 2019; Stock & Estrov, 2022b; van der Velden et al., 2003). Reverse transcriptase PCR-based MRD can also be used to detect cancer-related transcripts, including E2A/PBX1, TEL/AML1, and BCR/ABL (Lee et al., 2003; Madzo et al., 2003; Stock & Estrov, 2022b).
Proprietary Testing
ClonoSEQ (Adaptive Biotechnologies, Seattle, WA) is a commercially available NGS-based assay intended to assess MRD in certain types of cancer, such as MM and ALL. This test identifies rearrangements in certain receptor gene sequences, representing the level of MRD in a patient. This test typically uses genomic DNA extracted from bone marrow but may use circulating tumor DNA (ctDNA) (Herrera et al., 2016). After testing, a report is provided which includes each nucleotide sequence identified for tracking residual disease, the amount of each identified marker (per million cells), and whether MRD is determined to be present in the sample (Adaptive Biotechnologies, 2024).
SignateraTM is an MRD assay that uses ctDNA to attempt to inform the likelihood of cancer relapse earlier than standard of care tools. Whole-exome sequencing of an individual’s tumor tissue is first performed, allowing the identification of clonal somatic mutations which are expected to be present in all cells for the individual’s specific tumor. The test is customized to an individual through selection of 16 clonal, single nucleotide variants (SNVs). After customization, a blood sample may be obtained from the individual, from which the 16 SNVs may be amplified and detected. An individual is considered MRD-positive when at least two SNVs from the set of 16 are detected. According to the test makers, “a positive Signatera™ result predicts relapse with overall positive predictive value more than 98%” (Natera, 2024).
NavDx is a blood test developed by Naveris that detects trace amounts of HPV-associated tumor DNA in the bloodstream, specifically targeting residual disease or early recurrence of HPV-driven oropharyngeal cancers. It detects and analyzes the tumor tissue modified viral (TTMV)-HPV DNA fragments released by dying cancer cells to provide a TTMV-Score. The test indicates cancer activity before it is visible through imaging or other diagnostics (NavDx, 2024).
Analytical Validity
The EuroFlow Consortium has reported on the analytical validity of the use of an 8-color mFC for MRD. Theunissen et al. (2017) reported on the use of this methodology for B-cell precursor (BCP) ALL in a multicenter study (319 patients). Using samples containing more than four million cells, they note concordant results in 93% of samples, and “[m]ost discordances were clarified upon high-throughput sequencing of antigen-receptor rearrangements and blind multicenter reanalysis of flow cytometric data, resulting in an unprecedented concordance of 98% (97% for samples with MRD < 0.01%). In conclusion, the fully standardized EuroFlow BCP-ALL MRD strategy is applicable in >98% of patients with sensitivities at least similar to RQ-PCR (≤ 10−5), if sufficient cells (> 4 × 106, preferably more) are evaluated” (Theunissen et al., 2017). Another study reports the use of next-generation flow cytometry (NGF) using an “optimized 2-tube 8-color antibody panel” in five cycles to further increase the sensitivity. The authors report “a higher sensitivity for NGF-MRD vs conventional 8-color flow-MRD -MRD-positive rate of 47 vs 34% (P = 0.003). Thus, 25% of patients classified as MRD-negative by conventional 8-color flow were MRD-positive by NGF, translating into a significantly longer progression-free survival for MRD-negative vs MRD-positive CR [complete response] patients by NGF (75% progression-free survival not reached vs 7 months; P = 0.02).” Another study using a single-tube 10-fluorochrome analysis NGF method of MRD in myeloma reports a five-fold increase over the target minimum of 5 X 106 white blood cells per acquisition (Royston et al., 2016).
The FDA included an assessment of the analytical validity of ClonoSEQ in their approval summary of a de novo request evaluation. A total of 23 patients with MM, 21 patients with ALL, and 22 patients with other lymphoid malignancies were included. The study tested three different volumes of DNA: 500ng, 2μg, and 20 μg. Six MRD levels were tested for each sample, corresponding to the following amounts of malignant cells: 2.14, 6.13, 21.44, 61.26, 214.40, and 612.56. The authors found the coefficients of variance (%CV) to range from 72% at 2.14 cells to 21% at 612.56 cells. The authors noted that this precision trend was predictable, as ClonoSEQ is dependent on the number of cells evaluated instead of the actual MRD frequency. Regarding DNA extraction reproducibility, all samples were found to pass the “pre-established acceptance criteria of ± 30% MRD frequency.” Regarding precision of the nucleotide/base cells, the authors created a set of “baseline calibrating clonotype nucleotide sequences.” From this set, replicates of each sample used to create the calibration sequence were created and the disagreement rate was identified. Out of 442.5 million nucleotides, ClonoSEQ was found to have a disagreement rate of 3.5 parts per million. The FDA notes a Phred Score of >30 is considered a “high-quality base call for NGS applications”; ClonoSEQ had a Phred Score of 44.5. When compared to multiparametic flow cytometry (mpFC), both ClonoSEQ and mpFC were tested at five dilutions (from 5x10-7 to 1x10-2)s and both techniques were found to be of similar accuracy at frequencies above 1 x 10-4 (FDA, 2018).
In 2020, the FDA made a substantially equivalent decision on ClonoSEQ, providing in their summary an assessment with further analytical validity. Here, they looked at patients with CLL, measuring MRD levels from gDNA samples extracted from either bone marrow (22 patients) or blood samples (15 patients). The study tested six MRD levels in three different volumes of DNA: 500ng, 2μg, and 20 μg. In bone marrow samples, precision ranged from 59% CV at 2.14 cells to 20% CV at 612.56 cells. In blood samples, precision ranged from 53% CV at 3.10 cells to 19% CV at 765.70 cells. The authors note that “like BMA [bone marrow aspirate], the precision of the clonoSEQ assay in CLL blood is largely dependent on the number of malignant cells that are being evaluated by the assay.” From these data and those presented in the 2018 de novo approval document, the indications for use were developed. The authors report that “the clonoSEQ Assay measures minimal residual disease (MRD) to monitor changes in burden of disease during and after treatment. The test is indicated for use by qualified healthcare professionals in accordance with professional guidelines for clinical decision-making and in conjunction with other clinicopathological features” (FDA, 2020).
Othman Al-Sawaf et al. (2020) analyzed the clonal growth patterns of patients treated with venetoclax-obinutuzumab therapy within the CLL14 trial. In this case, MRD was analyzed using NGS via the adaptive clonoSEQ assay with cutoffs of 10-4, 10-5, and 10-6; “the limit-of-quantification of the clonoSEQ assay is less than 10-6.” A total of 432 patients with untreated CLL were either treated with chlorambucil or venetoclax, in combination with 7binutuzumab for the first six cycles. Using samples from peripheral blood (PB) collected every three to six months until nine years from last patient enrollment, the researchers found that two months post-treatment completion, among patients treated with venetoclax and obinutuzmab, 40% had uMRD levels < 10-6, 26% had uMRD levels that were ≥ 10-6 and < 10-5, 8% had uMRD levels ≥ 10-5 and < 10-4, 5% had uMRD levels ≥ 10-4 and < 10-2, and 3% had uMRD levels ≥ 10-2. In comparison, among patients treated with chlorambucil and obinutuzmab, 7% had uMRD levels < 10-6, 13% had uMRD levels ≥ 10-6 and < 10-5, 14% had uMRD levels ≥ 10-5 and < 10-4, 21% had uMRD levels ≥ 10-4 and < 10-2, and 26% had uMRD levels ≥ 10-2. Furthermore, “In a PFS landmark analysis after [end of treatment], patients in the [venetoclax-obinutuzumab] arm with MRD levels ≤ 10-5 had a 2-year PFS after [end of treatment] of approximately 93%, while patients with detectable MRD > 10-2 had a 2-year PFS of [approximately] 37%.” The average growth rate among patients treated with venetoclax-obinutuzumab was lower compared to the contrasting arm, and thus had a larger MRD doubling time. Because the clonal growth rate was lower, the study, using NGS, indicated “more effective MRD eradication and clonal growth modulation” (Othman Al-Sawaf et al., 2020; O. Al-Sawaf et al., 2020).
Nguyen Hoang et al. (2024) studied the analytical validity and clinical utilization of K-4CARE™, a comprehensive genomic profiling assay that integrates ctDNA tracking for residual cancer surveillance. K-4CARE™ is proposed to be a more time-efficient and tissue-efficient method than single or small gene panel testing. The assay tests for 473 cancer-relevant genes with a total length of 1.7 Mb. The authors rested the assay with 155 samples from 10 cancer types. “For detection of somatic SNVs and Indels, gene fusion and amplification, the assay had sensitivity of > 99%, 94% and >99% respectively, and specificity of > 99%.” Furthermore, “when CGP-informed mutations were used to personalize ctDNA tracking, the detection rate of ctDNA in liquid biopsy was 79%, and clinical utility in cancer surveillance was demonstrated in 2 case studies.” The authors concluded that the K-4CARE™ assay is “comprehensive and reliable” as a biomarker test for targeted therapy and immunotherapy. The authors also state that “integration of ctDNA tracking helps clinicians to further monitor treatment response and ultimately provide well-rounded care to cancer patients” (Nguyen Hoang et al., 2024).
Gunning et al. (2023) investigated the DNA stability and the ability to detect TTMV-HPV DNA analytes from patient samples stored in Streck collection tubes over a seven day interval. The ESR1 gene was chosen as the analyte for DNA stability analysis; DNA recovery was reported as the average, minimum, and maximum detected ESR1 per mL of plasma isolated. TTMV-HPV analyte detection was accomplished by measuring the TTMV-HPV DNA percent positivity rate, tracked daily. Percent positivity for TTMV-HPV DNA remained consistently between 26.2% – 32.5% from days one to seven. The average ESR1 values trended upward and more than doubled between days 1-6, which may be due to an increase in cellular lysis and DNA release within the sample over time. The LODs (limit of detection) for TTMV-HPV16, 18, 31, 33 and 35 DNA were reported to be: 0.56, 1.31, 0.63, 1.10 and 0.57 copies per microliter (copies/µL); LOQs (limit of quantitation) were reported as < 1.20, 3.56, 4.11, 4.00, and 3.50 copies/ µL. The analytical specificity of this study was reported to be between 0 and 0.32 copies/µL for the five high-risk HPV subtypes. The authors investigated “the ability of the NavDx test to obtain a response that is directly proportional to the concentration of analyte standard.” It was determined that all five HPV serotypes demonstrated linearity down to the limit of quantitation. The coefficients of determination (R2) equaled one for all serotypes at all timepoints. Intra-assay and inter-assay precision were investigated through determination of % CVs (coefficients of variation) of the measured concentrations of each HPV serotype:
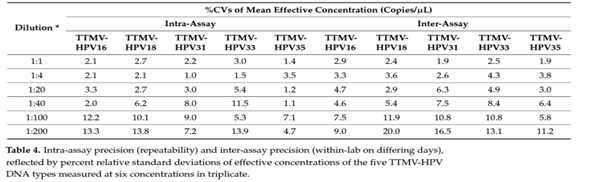
Table borrowed from Gunning et al. (2023).
Clinical Utility and Validity
The FDA de novo approval document for ClonoSEQ contains two clinical validation studies. The first study for MM in ClonoSEQ’s de novo approval document (DFCI 10-106) included 323 patients. The authors intended “to assess the ability of clonoSEQ to predict progression-free survival (PFS) and disease-free survival (DFS).” At the time of first MRD measurement, ClonoSEQ was found to be predictive of PFS at the MRD threshold of 10-5. Each 10-fold increase in MRD level was associated with a 70% increase in “event” rate across all MRD values (FDA, 2018).
A second clinical validation study described in the FDA’s de novo approval document was for ALL (AALL0232, AALL0331). A total of 273 samples were included (210 MRD ≤ 10-4, [negative], 63 MRD > 10-4 [positive]). The authors report that clonoSEQ MRD-negativity status was found to predict event-free survival (EFS) at all ages. MRD-positivity status was also associated with a 2.74-fold higher event risk compared to MRD-negativity status. Across all MRD values, a 10-fold increase in clonoSEQ MRD measurement was associated with a 50% increase in event rate and MRD-negative patients were found to have longer EFS compared to patients with higher frequencies of malignancies (FDA, 2018).
The FDA’s 2020 substantially equivalent approval document for ClonoSEQ analyzed two separate studies to “support that MRD as estimated with the clonoSEQ Assay is prognostic of patient outcomes in CLL …” The first study was for CLL (NCT02242942) and included 337 patients to evaluate the ability of ClonoSEQ to predict PFS. Samples were collected three months or later following treatment (FUM3) and MRD positivity was defined as > 1 x 10-5 [malignant cells]. Patients found to be MRD-positive had an “event risk” 6.64 times higher than the MRD-negative cohort. A 10-fold increase in MRD was also associated with a 2.35-fold increase in event risk. The authors also analyzed the results for other confounding factors and found “that the MRD level at FUM3 is a stronger predictor of PFS than age, sex, geographic region, Binet stage, or treatment arm of the clinical trial. Together, these results demonstrate the clinical validity of MRD measurement in CLL.” This study also found that “patients with clonoSEQ MRD ≤ 10-6 or between 10-6 and 10-5 had longer PFS, followed by patients with MRD between 10-5 and 10-4 and patients with MRD ≥ 10-4 (log-rank P = 4.902 x 10-31, Figure 11). These data demonstrate that patients with MRD ≤ 10-5 have better outcomes than patients with MRD > 10-5, and that increasing MRD levels above 10-5 are associated with an increased risk of progression within the follow-up time of this study” (FDA, 2020).
The second clinical validation described in FDA’s 2020 substantially equivalent approval document was also for CLL (NCT00759798). This study was a “phase 2 clinical trial that evaluated six cycles of fludarabine, cyclophosphamide, and rituximab (FCR) in 111 front-line chronic lymphocytic leukemia (CLL) patients with clonoSEQ ID samples and a corresponding 137 clonoSEQ MRD samples also evaluated by 4-color flow cytometry at an MRD threshold of 10-4 (NCT00759798) and with pertinent co-variate data. Within this cohort of 111 patients with flow MRD results, bone marrow was available for 75 patients and blood was available for 62 patients, of which 26 patients provided both blood and bone marrow. Due to some missing clinical covariates, three patients that provided bone marrow only, were excluded from analyses requiring these covariates. There was an association between PFS and continuous clonoSEQ MRD measurement in both blood and bone marrow, after end of treatment, where PFS is defined as the time from start of treatment until death, disease progression, or last time of disease assessment (p = 9.66 x 10-4 for blood, p = 2.13 x 10-4 for bone marrow). Additionally, patients who were MRD negative at a threshold ≤ 10-5 had superior progression-free survival compared to patients with MRD > 10-5 (p = .02 for blood and p = 8.17 x 10-5 for bone marrow… Taken together these results support the use of the clonoSEQ assay in CLL patients” (FDA, 2020).
Hay et al. (2019) evaluated the impact of MRD negativity status on relapse rates of ALL patients that post-chimeric antigen receptor [CAR] T-cell therapy. Per flow cytometry, 45 of 53 patients achieved an MRD-negative status. At a median follow-up of 30.9 months, the authors found that EFS and overall survival (OS) were significantly better in patients achieving MRD-negativity than patients that did not (median EFS: 7.6 months vs 0.8 months; median OS: 20 months vs five months). The authors also identified that the cytometric absence of the IGH index malignant clone was associated with better EFS (Hay et al., 2019).
Herrera et al. (2016) evaluated “whether the presence of ctDNA [circulating tumor DNA, measured with next-generation sequencing] was associated with outcome after allogeneic haematopoietic stem cell transplantation (HSCT) in lymphoma patients.” A total of 88 patients were included from a “phase 3 clinical trial of reduced-intensity conditioning HSCT in lymphoma.” Patients with detectable ctDNA three months after HSCT were found to have inferior progression-free survival compared to patients without detectable ctDNA (58% vs 84%, 2-year PFS rate). Detectable ctDNA was confered a 10.8-times higher risk of relapse/progression and a 3.9-times higher risk of progression/death compared to the non-detectable ctDNA group. The authors concluded that “detectable ctDNA is associated with an increased risk of relapse/progression, but further validation studies are necessary to confirm these findings and determine the clinical utility of NGS-based minimal residual disease monitoring in lymphoma patients after HSCT” (Herrera et al., 2016).
Perrot et al. (2018) examined the prognostic value of MRD (measured with NGS) in MM cases. A total of 127 patients achieved MRD negativity (defined as “the absence of tumor plasma cell within 1 000 000 bone marrow cells (< 10-6) at least once during maintenance therapy. At the start of therapy, MRD was found to be a strong prognostic factor for both progression-free survival as well as overall survival (hazard ratio = .22 and .24 respectively). From a previous cohort, the authors identified 233 patients labeled as MRD-negative, of which 120 were confirmed as MRD-negative with NGS (52%)” (Perrot et al., 2018).
Friend et al. (2020) investigated the impact of NGS-MRD in predicting relapse in ALL patients. Total body irradiation (TBI)-based regimens were the standard of care for ALL patients requiring allogeneic HSCT, but this procedure has numerous harmful side effects. The authors hypothesized that identifying MRD-negative patients may help some individuals avoid exposure to this radiation. The authors examined outcomes of 57 patients that received TBI and non-TBI regimens and found that relapse rates were similar for both methods of treatment. However, NGS-MRD positivity prior to treatment was “highly” predictive of relapse (for up to three years post-transplant). Based on their data, the authors suggested “that the decision to use either a TBI or non-TBI regimens in ALL should depend on NGS-MRD status, with conditioning regimens based on TBI reserved for patients that cannot achieve NGS-MRD negativity prior to allogeneic HSCT” (Friend et al., 2020).
Thörn et al. (2011) performed a comparative analysis of MFC and real-time quantitative polymerase chain reaction (RT-qPCR)-based MRD in pediatric ALL. The study, consisting of 726 follow-up samples from 228 children, used an MRD threshold of 0.1% and reported at day 29 a 84% concordance between the two different methods. For B-cell precursor ALL, the authors note that MFC was better at discriminating higher risk of bone marrow relapse (BMR), whereas RT-qPCR performed better for T-ALL. Regardless, the authors state, “MRD levels of ≥ 0.1%, detected by either method at day 29, could not predict isolated extramedullary relapse.” They conclude that “both methods are valuable clinical tools for identifying childhood ALL cases with increased risk of BMR” (Thörn et al., 2011).
Wood et al. (2018) compared high-throughput sequencing (HTS) of IGH and TRG genes to flow cytometry (FC) to evaluate measurable residual disease detection at the end of induction chemotherapy in pediatric patients with newly diagnosed B-ALL [B-cell acute lymphoblastic leukemia]. A total of 619 paired pre-treatment and end of induction bone marrow samples were included. At an MRD threshold of 0.01%, both HTS and FC showed similar event-free survival (EFS) and OS for both MRD-positive and MRD-negative patients. However, HTS identified 55 more patients as “MRD-positive” compared to FC. These “discrepant” patients were found to have worse outcomes than FC MRD-negative patients. HTS was also found to identify 19.9% of “standard risk” (SR) without MRD at any detectable level with excellent EFS and OS (98.1% and 100% respectively). The authors suggested that “the higher analytic sensitivity and lower false-negative rate of HTS improves upon FC for MRD detection in pediatric B-ALL by identifying a novel subset of patients at end of induction who are essentially cured using current chemotherapy and identifying MRD at 0.01% in up to one-third of patients who are missed at the same threshold by FC” (Wood et al., 2018).
Rawstron et al. (2016) conducted a parallel analysis of MRD using both ClonoSEQ and multiparameter flow cytometry in CLL as part of the European Research Initiative on CLL (ERIC) study. The MFC approach used within the ERIC study is validated to the level of 10-5 and consists of six different markers — CD5, CD19, CD20, CD43, CD49b, and CD81. The ERIC study reports that the ClonoSEQ method “provides good linearity to a detection limit of one in a million (10-6).” The authors also note, “a parallel analysis of high-throughput sequencing using the ClonoSEQ assay showed good concordance with flow cytometry results at the 0.010% (10-4) level, the MRD threshold defined in the 2008 International Workshop on CLL guidelines… The combination of both technologies would permit a highly sensitive approach to MRD detection while providing a reproducible and broadly accessible method to quantify residual disease and optimize treatment in CLL” (Rawstron et al., 2016).
Thompson et al. (2019) evaluated 62 patients with CLL that were considered negative for MRD by flow cytometry (sensitivity of 10-4). Using ClonoSEQ, the authors found that 72.6% of these MRD-negative patients were MRD-positive by ClonoSEQ (a discordant result). Only 27.4% of patients were found to be negative by both methods. The authors also found that patients that were negative by both methods were found to have superior PFS compared to patients that were only negative by flow cytometry, thereby suggesting that ClonoSEQ was a superior prognostic discriminator (Thompson et al., 2019).
Z. Wang et al. (2019) published a study on the applicability of multiparameter (multicolor) flow cytometry (MFC) for detecting MRD to predict relapse in patients with AML after allogeneic transplantation. The researchers determined MFC and MRD status using real-time quantitative polymerase chain reaction (RT-qPCR) from 158 bone marrow samples from 44 different individuals and compared the statuses between the two. They noted that “Strong concordance was found between MFC-based and RT-qPCR-based MRD status (κ = 0.868).” Moreover, for individuals in complete remission (CR), “the positive MRD status detected using MFC was correlated with a worse prognosis [HRs (P values) for relapse, event-free survival, and overall survival: 4.83 (<0.001), 2.23 (0.003), and 1.79 (0.049), respectively]; the prognosis was similar to patients with an active disease before HSCT [hematopoietic stem cell transplantation]” (Z. Wang et al., 2019).
Carlson et al. (2019) published a cost-effectiveness study of NGS-based MRD testing during maintenance treatment for MM. The authors compared use of MRD testing to no MRD testing. A Markov model with six health states was developed; “MRD positive or MRD negative, on or off treatment, relapsed, or dead.” From there, the authors compared yearly NGS-MRD to no MRD testing over a lifetime horizon. Overall, the authors found that “MRD testing saved $1,156,600 over patients remaining lifetime.” Health outcomes were found to slightly favor MRD testing (0.01 quality-adjusted life years [QALYs]) compared to no testing. The authors concluded that “NGS MRD testing is cost saving, with potential QALY gains due to avoidance of [treatment-related adverse events] compared with no testing for MM patients on maintenance therapy” (Carlson et al., 2019).
Medina et al. (2020) evaluated MRD three months after transplantation in 106 myeloma patients, noting that “detecting persistent minimal residual disease (MRD) allows the identification of patients with an increased risk of relapse and death.” In this study, they compared the results of NGS with NGF, where they noted that “correlation between NGS and NGF was high (R2 = 0.905). The three-year PFS rates by NGS and NGF were longer for undetectable vs. positive patients (NGS: 88.7% vs. 56.6%; NGF: 91.4% vs. 50%; p < 0.001 for both comparisons), which resulted in a three-year overall survival (OS) advantage (NGS: 96.2% vs. 77.3%; NGF: 96.6% vs. 74.9%, p < 0.01 for both comparisons). In the Cox regression model, NGS and NGF negativity had similar results but favoring the latter in PFS (HR: 0.20, 95% CI: 0.09-0.45, p < 0.001) and OS (HR: 0.21, 95% CI: 0.06-0.75, p = 0.02). All these results reinforce the role of MRD detection by different strategies in patient prognosis and highlight the use of MRD as an endpoint for multiple myeloma treatment” (Medina et al., 2020).
Goicoechea et al. (2021) examined MRD as a possible endpoint marker in MM. They note that while patients with MM that carry standard- or high-risk cytogenetic abnormalities (CA) are achieving similar CR rates, high-risk patients have an inferior PFS. They note that this “questions the legitimacy of CR as a treatment endpoint ...” Using NGF cytometry to evaluate MRD in MM patients, they compared standard- vs high-risk CAs (n = 300 and 90, respectively) and identified mechanisms that determine MRD resistance in both patient subgroups (n = 40). In patients achieving undetectable MRD with either standard- or high-risk CAs, the 36-month PFS rates were higher than 90%. In comparison, patients with persistent MRD had a median PFS of about three (standard-risk CA) and two (high-risk CA) years. They found that “further use of NGF to isolate MRD, followed by whole-exome sequencing of paired diagnostic and MRD tumor cells, revealed greater clonal selection in patients with standard-risk CAs, higher genomic instability with acquisition of new mutations in high-risk MM, and no unifying genetic event driving MRD resistance.” Ultimately, their results support “undetectable MRD as a treatment endpoint for patients with MM who have high-risk CAs and proposes characterizing MRD clones to understand and overcome MRD resistance” (Goicoechea et al., 2021).
Subhash et al. (2022) investigated the “feasibility of using whole-genome sequencing (WGS) data to design tumour-specific polymerase chain reaction (PCR)-based MRD tests (WGS-MRD)” for children with cancers with high-risk of relapse, like ALL, high-risk neuroblastoma (HR-NB), and Ewing sarcoma (EWS). The significance of this study was that pediatric solid tumor MRD DNA-based assays currently remains experimental, and remains focused on mRNA, methylated DNA, or microRNA. Through their experimentation, the researchers found that “sensitive WGS-MRD assays were generated for each patient and allowed quantification of 1 tumour cell per 10-4 (0.01%) – 10-5 (0.001%) mononuclear cells.” They also found that WGS-MRD and Ig/TCR-MRD assays were concordant in ALL, and the WGS-MRD assays “showed good concordance between quantitative PCR and droplet digital PCR formats.” The WGS-MRD assay clinical samples also correlated with disease course and was found to be more sensitive than RNA-based MRD assays. This demonstrated how WGS could aid the development of MRD assays for pediatric cancers as it has been done in adults (Subhash et al., 2022).
Kater et al. (2019) utilized the MURANO study to investigate the effects of a fixed duration treatment with venetoclax and ritixumab on MRD. The study itself demonstrated “significant progression-free survival (PFS) benefit for fixed-duration venetoclax-rituximab compared with bendamustine-rituximab in relapsed/refractory chronic lymphocytic leukemia.” However, among patients who received venetoclax-rituximab, there was a “higher rate of PB [peripheral blood] undetectable MRD (uMRD; less than 10-4) at EOCT [end of combination therapy] (62% v 13%) with superiority sustained through month 24 (end of therapy),” which predicted longer PFS. At the end of the therapy treatment period, 70% of patients remained in uMRD and 98% without disease progression. This demonstrated the correlation between uMRD and PFS using PB MRD in the setting of venetoclax-combination treatment (Kater et al., 2019).
Kim et al. (2022) investigated the clinical utility of NGF-based MRD assessment in a heterogeneous population of patients with MM at the Samsung Medical Center in Korea and found that “sCR samples showed a lower MRD-positive rate (25%) than CR (43%) and VGPR (53%) samples, although the difference was not significant (P = 0.051).” When evaluating survival analysis based on clinical response and MRD, “PFS in VGPR patients was lower than that in sCR/CR patients (P < 0.001) …whereas PFS in VGPR patients was lower than that in sCR/CR patients (P<0.001).” Cytogenetic risk survival analysis yielded similar results: “There was no significant difference in PFS between patients with high-risk and standard-risk cytogenetics (P = 0.222)” and “Further analysis according to MRD status also revealed no significant difference in PFS in patients with standard-risk cytogenetics (P = 0.246),” though it is worth noting that “among patients with high-risk cytogenetics, MRD-positive patients showed lower PFS than MRD-negative patients (P = 0.016).” Overall, the authors concluded that “Sustained MRD negativity was only observed in patients with sustained sCR, and their PFS was superior to that of patients who were not MRD-negative (P = 0.035),” though by their own admission the study was limited by the sample size (n = 12 over 18 months) (Kim et al., 2022).
There is a growing pool of literature investigating the use of ctDNA-based MRD detection in the clinical context of solid tumors. Some studies suggest that inclusion of MRD assessment may offer some improvement over standard of care; this has been reported for gastrointestinal malignancies (Zhang et al., 2021), skin cancer (Eroglu et al., 2023; Khaddour et al., 2022), lung cancer (Zhong et al., 2023), and especially colorectal cancer, where several reports demonstrate potential benefits of ctDNA measurement, which include response monitoring, prognosis, post-surgical surveillance, chemotherapeutic management, and informing recurrence risk (Hofste et al., 2023; Sato et al., 2023; Tie et al., 2019; Tie et al., 2016; Y. Wang et al., 2019).
SignateraTM has been clinically validated across several cancer types for recurrence monitoring. In the context of colorectal cancer, Loupakis et al. (2021) demonstrated that a two-timepoint analysis of ctDNA (a baseline measure plus last follow-up time point–either the time of radiologic progression or last evidence of radiologic disease's absence) improved the sensitivity of the assay. For patients who were ctDNA positive at both timepoints, the authors reported that the assay demonstrated a sensitivity of 91.4%, specificity of 93.3%, and positive predictive value (PPV) of 96.7%. Reinert et al. (2019) also investigated the validity of ctDNA for MRD detection for individuals with stages I-III colorectal cancer and reported promising metrics; the test demonstrated a sensitivity of 88%, specificity of 98%, PPV of 93.3%, and negative predictive value (NPV) of 96.7%. However, Fakih et al. (2022) recently determined that SignateraTM may not “provide advantages as a surveillance strategy compared with standard imaging combined with CEA levels when performed per National Comprehensive Cancer Network guidelines,” after a cohort study failed to demonstrate a sufficient ability (sensitivity) of the test to detect disease recurrence in individuals with resected colorectal cancer.
Coombes et al. (2019) described the ability of ctDNA to detect disease recurrence ahead of clinical or radiological approaches for individuals with breast cancer with a sensitivity of 89%, specificity of 100%, PPV of 100%, and NPV of 94%. Christensen et al. (2019) found that ctDNA accurately identified individuals diagnosed with advanced bladder cancer who eventually relapsed (sensitivity of 100% and specificity of 98%). For individuals with non-small cell lung cancer, Abbosh et al. (2017) reported that SignateraTM detected relapse with 93% sensitivity and 90% specificity.
While evidence supporting MRD analysis for solid tumors continues to emerge, a consensus on the clinical validity and utility of this approach has not yet been reached. Sullivan et al. (2023) cite several limitations of ctDNA-based approaches to the management of gastrointestinal cancer; O'Sullivan et al. (2023) argue that the utility of this technology in the context of non-small cell lung cancer is still investigational; Moding et al. (2021) cite a lack of prospective clinical trials confirming clinical utility of ctDNA MRD assessment in general; and, Jacome and Johnson (2023) argue that there is not yet consensus on how to apply the results of ctDNA-based testing in the management of colorectal cancer.
Faulkner et al. (2023) conducted a systematic review and meta-analysis to study the utility of ctDNA for detecting MRD in colorectal cancer. The review included 37 studies involving 3002 patients. The authors calculated hazard ratios and PFS based on ctDNA detection. Overall, there was a “poorer” PFS associated with ctDNA detection at the first liquid biopsy post-surgery, with a hazard ratio of 6.92. Further, “this effect was also seen in subgroup analysis by disease extent, adjuvant chemotherapy and assay type.” The authors conclude that “ctDNA detection post-surgery is associated with a greater propensity to disease relapse and is an independent indicator of poor prognosis,” but note that “prior to incorporation into clinical practice, consensus around timing of measurements and assay methodology are critical” (Faulkner et al., 2023).
Berger et al. (2022) evaluated the use of circulating tumor tissue modified viral (TTMV)-HPV DNA as a biomarker for detecting recurrence in patients with HPV-driven oropharyngeal squamous cell carcinoma (OPSCC). The authors conducted a retrospective analysis of 1,076 patients from 108 U.S sites who were at least three months post-treatment and had undergone TTMV-HPV DNA testing (NavDx) during routine surveillance between February 2020 and June 2021. The results determined that 80 out of 1,076 (7.4%) tested positive for TTMV-HPV DNA. Among these, 21 patients had known recurrences at the time of the first positive test, while 59 did not, and 93% of the latter group subsequently developed confirmed recurrences. The overall positive predictive value for the TTMV-HPV DNA test was 95%, and the negative predictive value was also 95%. These findings suggest that TTMV-HPV DNA testing can serve as an effective surveillance tool, often identifying recurrences earlier than traditional clinical evaluations and imaging techniques, and may influence future surveillance strategies for HPV-related cancers (Berger et al., 2022).
National Comprehensive Cancer Network (NCCN)
The NCCN has published several relevant guidelines on management of MRD in hematologic malignancies.
Multiple Myeloma (MM)
For MM, MRD is considered an “important” prognostic factor. The NCCN recommends measuring MRD during follow-up/surveillance for symptomatic MM patients after response to primary therapy “as indicated for prognostication.” Next-generation flow and next-generation sequencing (or both) are recommended for methodology and a sensitivity of 1 in 105 (or better) is recommended for accuracy. The NCCN recommends to “consider baseline clone identification and storage of aspirate sample for future minimal residual disease (MRD) testing by NGS.” MRD is a required criterion listed within the IMWG MM response criteria. The NCCN notes that “for MRD there is no need for two consecutive assessments, but information on MRD after each treatment state is recommended (eg, after induction, high-dose therapy/Autologous stem cell transplant (ASCT), consolidation, maintenance). MRD tests should be initiated only at the time of suspected complete response.” Sustained MRD-negative status is only confirmed when taken a minimum of one year apart, but “subsequent evaluations can be used to further specify the duration of negativity (eg, MRD-negative at 5 years).” The NCCN also notes that MRD is being used in post-hematopoietic stem cell transplantation treatment assessments (NCCN, 2024e).
Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma
The NCCN remarks that “undetectable MRD in the peripheral blood at the end of fixed duration treatment is an important predictor of efficacy.” The NCCN recommends performing MRD assessment with an assay at a sensitivity of 10-4 “according to the standardized European Research Initiative on CLL (ERIC) method or standardized NGS method.” NCCN also stated that “allele-specific oligonucleotide polymerase chain reaction (ASO-PCR) and six-color flow cytometry (MRD flow) are the two validated methods used for the detection of MRD at the level of 10-4 to 10-5. NGS-based assays have been shown to be more sensitive, thus allowing for the detection of MRD at the level of 10-6“ (NCCN, 2024c).
Acute Myeloid Leukemia (AML)
The NCCN recommends measuring MRD “upon completion of initial induction” and “before allogenic HCT.” The NCCN also states, “Additional time points should be guided by the regimen used.” NGS-based assays to detect mutated genes are not routinely used in AML, as the sensitivity of PCR-based assays and flow cytometry is superior to what is achieved by NGS. The NCCN states that “if using flow cytometry to assess MRD, it is recommended that a specific MRD assay is utilized, but, most importantly, that it is interpreted by an experienced hematopathologist” since there are differences between “diagnostic threshold assessments and MRD assessments.” They also note that “some evidence suggest MRD testing may be more prognostic than KIT mutation status in CBF-AML, but this determination depends on the method used to assess MRD and the trend of detectable MRD.” The NCCN further states that “Based on the techniques, the optimal sample for MRD assessment is either peripheral blood (NPM1 PCR-based techniques) or an early, dedicated pull of the BM aspirate (i.e., other PCR, flow cytometry, NGS). The quality of the sample is of paramount importance to have reliable evaluation” (NCCN, 2024b).
Acute Lymphoblastic Leukemia (ALL)/Pediatric Acute Lymphoblastic Leukemia
The NCCN states that MRD quantification is an “essential component of patient evaluation over the course of sequential ALL therapy,” noting the prognostic significance of MRD. Techniques used to assess for MRD include: flow cytometry assays, real-time quantitative PCR assays, reverse transcriptase quantitative PCR assays, and NGS-based assays. NGS is recognized as one of the most sensitive methods at detection levels of 10-6, as can some PCRs. An entire section within the ALL guidelines is devoted to MRD assessment. They note the timing of MRD assessment to be as follows:
- Upon completion of initial induction.
- End of consolidation.
- Additional time points should be guided by the regimen used and risk features.
- Serial monitoring frequency may be increased in patients with molecular relapse or persistent low-level disease burden.
- For some techniques, a baseline sample (i.e., prior to treatment) is needed to characterize the leukemic clone for subsequent MRD assessment.
The guidelines also recommend “For patients with negative MRD by flow cytometry but positive MRD by an FDA-approved NGS assay, consider repeat testing before consolidation is started to confirm MRD status” (NCCN, 2024a). Overall, MRD has a strong correlation with risks for relapse and is considered to have a high prognostic value. MRD has a role in identifying optimal treatments for patients, both adult and pediatric, with ALL (NCCN, 2024a, 2024f).
Hairy Cell Leukemia
The NCCN writes that, for a complete response (CR) with or without MRD, “if CR is achieved, an IHC [immunohistochemistry] assessment of the percentage of MRD will be useful to stratify patients based on level of CR (with or without evidence of MRD)”. The NCCN notes that “it has been suggested that MRD monitoring as a component of response assessment should be incorporated in all clinical trials for relapsed HCL” but clarifies that “MRD assessment is not recommended (outside of clinical trials) as part of response evaluation” (NCCN, 2024d).
International Myeloma Working Group (IMWG)
The IMWG recommends assessing MRD response at a sensitivity of 1/105 nucleated cells or better (Kumar et al., 2016).
European Myeloma Network (EMN)
Regarding NGS in assessment of MRD in MM, the EMN writes that “Results from next-generation sequencing are highly concordant with flow-based MRD detection, highly reproducible and reach a sensitivity of 10−6” and that the primary restraints for NGS are “a lack of standardization and limited commercial availability” (Caers et al., 2018).
American Society of Clinical Oncology (ASCO) and Cancer Care Ontario (CCO)
These joint guidelines focus on treatment of MM. Their MRD-related recommendations are listed below:
- “There is insufficient evidence to make modifications to maintenance therapy based on depth of response, including MRD status.”
- “MRD-negative status has been associated with improved outcomes, but it should not be used to guide treatment goals outside the context of a clinical trial.”
- “There is insufficient evidence to support change in type and length of therapy based on depth of response as measured by conventional IMWG approaches or MRD.”
- “There are not enough data to recommend risk-based versus response-based duration of treatment (such as MRD)” (Mikhael et al., 2019).
The ASCO also fully endorsed the “Initial Diagnostic Work-Up of Acute Leukemia” released jointly by the College of American Pathologists and the American Society of Hematology in 2018 (de Haas et al., 2018).
European LeukemiaNet (ELN) MRD Working Party
The ELN states, “Measurable residual disease (MRD; previously termed minimal residual disease) is an independent, postdiagnosis, prognostic indicator in acute myeloid leukemia (AML) that is important for risk stratification and treatment planning, in conjunction with other well-established clinical, cytogenetic, and molecular data assessed at diagnosis.” The ELN remarks that quantitative PCR is applicable to approximately 40% of AML patients with “1 or more suitable abnormalities.” However, NGS for MRD assessment may provide assessment to an additional 40% – 50% of AML patients, as NGS can “theoretically, be applied to all leukemia-specific genetic aberrations.” The ELN recommends a sensitivity of at least 1 / 103 cells, and states that NGS platforms will be used after careful validation (Schuurhuis et al., 2018).
International Workshop on Chronic Lymphocytic Leukemia (iwCLL)
The iwCLL published guidelines on CLL in 2018. In it, they consider MRD assessment to be a necessary component in identifying complete remission of CLL. The iwCLL also writes that eradication of leukemia is a “desired end point.” They go on to state: “Use of sensitive multicolor flow cytometry, PCR, or next-generation sequencing can detect MRD in many patients who achieved a complete clinical response. … Six-color flow cytometry (MRD flow), allele-specific oligonucleotide PCR, or high-throughput sequencing using the ClonoSEQ assay are reliably sensitive down to a level of < 1 CLL cell in 10 000 leukocytes” (Hallek et al., 2018).
College of American Pathologists (CAP) and the American Society of Hematology (ASH)
This CAP/ASH joint guideline was published in 2017 and focuses on Initial Diagnostic Workup of Acute Leukemia (AL). The guideline strongly recommends that “For patients with suspected or confirmed AL, the pathologist or treating clinician should ensure that flow cytometry analysis or molecular characterization is comprehensive enough to allow subsequent detection of MRD.” The guideline also notes that MRD is a “powerful” predictor of adverse outcome in patients with AL (Arber et al., 2017).
This guideline was endorsed by the American Society of Clinical Oncology in 2018 (Arber et al., 2017).
European Society for Medical Oncology (ESMO)
Chronic Lymphocytic Leukaemia
The ESMO notes that “detection of MRD by multicolour flow cytometry or RT-PCR has a strong prognostic impact following CIT73,74 as well as venetoclax plus CD20-antibody combinations. A total of 75 patients with undetectable MRD after therapy show a longer response duration and survival. Additional clinical consequences of MRD positivity after therapy with respect to treatment escalation remain unclear… Therefore, MRD assessment is not generally recommended for monitoring after therapy outside clinical studies. This may change soon, as increasing efforts are made to determine whether therapy with targeted agents could be discontinued on the basis of MRD status” (B. Eichhorst et al., 2021). The guidelines are also endorsed by the European Hematology Association (Barbara Eichhorst et al., 2021).
Acute Lymphoblastic Leukaemia
The ESMO writes that “Quantification of MRD is a major and well-established risk factor and should be obtained whenever possible for all patients also outside of clinical trials…If MRD is measured by flow cytometry, a good MRD response is often defined as less than 10−3 , although MRD levels less than 10−4 can be achieved with the 8–12 colour flow cytometers” (Hoelzer et al., 2016).
Multiple Myeloma
The ESMO states, “One of the most significant improvements in the response criteria is the introduction of minimal residual disease (MRD) both in the bone marrow (BM) [using either next-generation sequencing or next-generation flow cytometry (NGF)] and outside the BM [using positron emission tomography-computed tomography (PET-CT); imaging MRD]. MRD negativity in the BM in patients who have achieved conventional complete response (CR) consistently correlates with prolonged progression-free survival (PFS) and overall survival (OS) in both newly diagnosed MM (NDMM) and relapsed/refractory MM (RRMM) patients.” ESMO also notes that “MRD has been found to be a surrogate endpoint for PFS in patients receiving first-line treatment. Therefore, MRD may be used as an endpoint to accelerate drug development. The use of MRD to drive treatment decisions is under investigation” (Dimopoulos et al., 2021).
Acute Myeloid Leukaemia
The ESMO includes MRD status as part of the treatment algorithm for AML. They state, “Morphological enumeration of the blast percentage should be refined by immunophenotypic or molecular MRD assessment in patients with < 10% blasts. ELN recommendations on MRD assessment in AML specify its clinical use and technical requirements. It is recommended to assess MRD by reverse transcriptase polymerase chain reaction (RT-PCR) for patients positive for NPM1mut, RUNX1-RUNX1T1, CBFB-MYH11 or PML-RARA fusion genes; ~ 40% of all AML patients. In the remaining patients, MRD should be assessed by MFC, which relies on antigens aberrantly expressed by leukaemic cells that can be found in > 90% of AML patients. Many clinical studies have shown the strong prognostic impact of MRD, as measured by MFC, with levels 0.1% defined as positive” (Heuser et al., 2020).
Hairy Cell Leukaemia
Concerning hairy cell leukemia, ESMO notes, “Recently, monoclonal antibodies that detect the mutated BRAF protein have been developed and shown to be useful for the diagnosis and detection of minimal residual disease (MRD).” Within the section on response evaluation, ESMO states, “Immunophenotypic analysis of peripheral blood or bone marrow is not required but is useful to detect MRD… The eradication of MRD is generally not recommended in routine clinical practice. Assessment of response should be performed 4 – 6 months after treatment with 2-CldA and after 8 – 9 courses of DCF. Relapse is defined as any deterioration in blood counts related to the detection of hairy cells in peripheral blood and/or bone marrow” (Robak et al., 2015).
References
- Abbosh, C., Birkbak, N. J., Wilson, G. A., Jamal-Hanjani, M., Constantin, T., Salari, R., Le Quesne, J., Moore, D. A., Veeriah, S., Rosenthal, R., Marafioti, T., Kirkizlar, E., Watkins, T. B. K., McGranahan, N., Ward, S., Martinson, L., Riley, J., Fraioli, F., Al Bakir, M., . . . Swanton, C. (2017). Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature, 545(7655), 446-451. https://doi.org/10.1038/nature22364
- Adaptive Biotechnologies. (2024). ClonoSEQ MRD Monitoring. Retrieved 07/10/2020 from https://www.clonoseq.com/
- Al-Sawaf, O., Zhang, C., Robrecht, S., Wilson, C., Tandon, M., Ching, T., Fink, A.-M., Ritgen, M., Tausch, E., Kreuzer, K.-A., Schary, W., Wendtner, C.-M., Eichhorst, B., Stilgenbauer, S., Jiang, Y., Hallek, M., & Fischer, K. (2020). Clonal Dynamics after Venetoclax-Obinutuzumab Therapy: Novel Insights from the Randomized, Phase 3 CLL14 Trial. Blood, 136(Supplement 1), 22-23. https://doi.org/10.1182/blood-2020-136977
- Al-Sawaf, O., Zhang, C., Tandon, M., Sinha, A., Fink, A. M., Robrecht, S., Samoylova, O., Liberati, A. M., Pinilla-Ibarz, J., Opat, S., Sivcheva, L., Le Dû, K., Fogliatto, L. M., Niemann, C. U., Weinkove, R., Robinson, S., Kipps, T. J., Tausch, E., Schary, W., . . . Fischer, K. (2020). Venetoclax plus obinutuzumab versus chlorambucil plus obinutuzumab for previously untreated chronic lymphocytic leukaemia (CLL14): follow-up results from a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol, 21(9), 1188-1200. https://doi.org/10.1016/s1470-2045(20)30443-5
- Arber, D. A., Borowitz, M. J., Cessna, M., Etzell, J., Foucar, K., Hasserjian, R. P., Rizzo, J. D., Theil, K., Wang, S. A., Smith, A. T., Rumble, R. B., Thomas, N. E., & Vardiman, J. W. (2017). Initial Diagnostic Workup of Acute Leukemia: Guideline From the College of American Pathologists and the American Society of Hematology. Arch Pathol Lab Med, 141(10), 1342-1393. https://doi.org/10.5858/arpa.2016-0504-CP
- Bai, Y., Orfao, A., & Chim, C. S. (2018). Molecular detection of minimal residual disease in multiple myeloma. Br J Haematol, 181(1), 11-26. https://doi.org/10.1111/bjh.15075
- Berger, B. M., Hanna, G. J., Posner, M. R., Genden, E. M., Lautersztain, J., Naber, S. P., Del Vecchio Fitz, C., & Kuperwasser, C. (2022). Detection of Occult Recurrence Using Circulating Tumor Tissue Modified Viral HPV DNA among Patients Treated for HPV-Driven Oropharyngeal Carcinoma. Clinical Cancer Research, 28(19), 4292-4301. https://doi.org/10.1158/1078-0432.ccr-22-0562
- Brüggemann, M., Raff, T., Flohr, T., Gökbuget, N., Nakao, M., Droese, J., Lüschen, S., Pott, C., Ritgen, M., Scheuring, U., Horst, H. A., Thiel, E., Hoelzer, D., Bartram, C. R., & Kneba, M. (2006). Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukemia. Blood, 107(3), 1116-1123. https://doi.org/10.1182/blood-2005-07-2708
- Caers, J., Garderet, L., Kortum, K. M., O'Dwyer, M. E., van de Donk, N., Binder, M., Dold, S. M., Gay, F., Corre, J., Beguin, Y., Ludwig, H., Larocca, A., Driessen, C., Dimopoulos, M. A., Boccadoro, M., Gramatzki, M., Zweegman, S., Einsele, H., Cavo, M., . . . Engelhardt, M. (2018). European Myeloma Network recommendations on tools for the diagnosis and monitoring of multiple myeloma: what to use and when. Haematologica, 103(11), 1772-1784. https://doi.org/10.3324/haematol.2018.189159
- Carlson, J. J., Eckert, B., & Zimmerman, M. (2019). Cost-effectiveness of next-generation sequencing minimal residual disease testing during maintenance treatment for multiple myeloma. Journal of Clinical Oncology, 37(15_suppl), e19529-e19529. https://doi.org/10.1200/JCO.2019.37.15_suppl.e19529
- Christensen, E., Birkenkamp-Demtroder, K., Sethi, H., Shchegrova, S., Salari, R., Nordentoft, I., Wu, H. T., Knudsen, M., Lamy, P., Lindskrog, S. V., Taber, A., Balcioglu, M., Vang, S., Assaf, Z., Sharma, S., Tin, A. S., Srinivasan, R., Hafez, D., Reinert, T., . . . Dyrskjot, L. (2019). Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma. J Clin Oncol, 37(18), 1547-1557. https://doi.org/10.1200/JCO.18.02052
- Coombes, R. C., Page, K., Salari, R., Hastings, R. K., Armstrong, A., Ahmed, S., Ali, S., Cleator, S., Kenny, L., Stebbing, J., Rutherford, M., Sethi, H., Boydell, A., Swenerton, R., Fernandez-Garcia, D., Gleason, K. L. T., Goddard, K., Guttery, D. S., Assaf, Z. J., . . . Shaw, J. A. (2019). Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin Cancer Res, 25(14), 4255-4263. https://doi.org/10.1158/1078-0432.CCR-18-3663
- de Haas, V., Ismaila, N., Advani, A., Arber, D. A., Dabney, R. S., Patel-Donnelly, D., Kitlas, E., Pieters, R., Pui, C.-H., Sweet, K., & Zhang, L. (2018). Initial Diagnostic Work-Up of Acute Leukemia: ASCO Clinical Practice Guideline Endorsement of the College of American Pathologists and American Society of Hematology Guideline. Journal of Clinical Oncology, 37(3), 239-253. https://doi.org/10.1200/JCO.18.01468
- Del Giudice, I., Raponi, S., Della Starza, I., De Propris, M. S., Cavalli, M., De Novi, L. A., Cappelli, L. V., Ilari, C., Cafforio, L., Guarini, A., & Foà, R. (2019). Minimal Residual Disease in Chronic Lymphocytic Leukemia: A New Goal? Front Oncol, 9, 689. https://doi.org/10.3389/fonc.2019.00689
- Dimopoulos, M. A., Moreau, P., Terpos, E., Mateos, M. V., Zweegman, S., Cook, G., Delforge, M., Hajek, R., Schjesvold, F., Cavo, M., Goldschmidt, H., Facon, T., Einsele, H., Boccadoro, M., San-Miguel, J., Sonneveld, P., Mey, U., guidelines@ehaweb.org, E. H. A. G. C. E. a., & clinicalguidelines@esmo.org, E. G. C. E. a. (2021). Multiple myeloma: EHA-ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up(dagger). Ann Oncol, 32(3), 309-322. https://doi.org/10.1016/j.annonc.2020.11.014
- Eichhorst, B., Ghia, P., & on behalf of the, E. H. A. G. C. (2021). EHA Endorsement of ESMO Clinical Practice Guidelines for Diagnosis, Treatment, and Follow-up of Chronic Lymphocytic Leukemia. HemaSphere, 5(1). https://doi.org/10.1097/HS9.0000000000000520
- Eichhorst, B., Robak, T., Montserrat, E., Ghia, P., Niemann, C. U., Kater, A. P., Gregor, M., Cymbalista, F., Buske, C., Hillmen, P., Hallek, M., Mey, U., & clinicalguidelines@esmo.org, E. G. C. E. a. (2021). Chronic lymphocytic leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol, 32(1), 23-33. https://doi.org/10.1016/j.annonc.2020.09.019
- Eroglu, Z., Krinshpun, S., Kalashnikova, E., Sudhaman, S., Ozturk Topcu, T., Nichols, M., Martin, J., Bui, K. M., Palsuledesai, C. C., Malhotra, M., Olshan, P., Markowitz, J., Khushalani, N. I., Tarhini, A. A., Messina, J. L., & Aleshin, A. (2023). Circulating tumor DNA-based molecular residual disease detection for treatment monitoring in advanced melanoma patients. Cancer, 129(11), 1723-1734. https://doi.org/10.1002/cncr.34716
- Fakih, M., Sandhu, J., Wang, C., Kim, J., Chen, Y. J., Lai, L., Melstrom, K., & Kaiser, A. (2022). Evaluation of Comparative Surveillance Strategies of Circulating Tumor DNA, Imaging, and Carcinoembryonic Antigen Levels in Patients With Resected Colorectal Cancer. JAMA Netw Open, 5(3), e221093. https://doi.org/10.1001/jamanetworkopen.2022.1093
- Faulkner, L. G., Howells, L. M., Pepper, C., Shaw, J. A., & Thomas, A. L. (2023). The utility of ctDNA in detecting minimal residual disease following curative surgery in colorectal cancer: a systematic review and meta-analysis. British journal of cancer, 128(2), 297-309.
- FDA. (2018, 09/28/2020). EVALUATION OF AUTOMATIC CLASS II DESIGNATION FOR clonoSEQ® ASSAY DECISION SUMMARY. Retrieved 07/13/2020 from https://www.accessdata.fda.gov/cdrh_docs/reviews/DEN170080.pdf
- FDA. (2020). 510(k) SUBSTANTIAL EQUIVALENCE DETERMINATION DECISION SUMMARY. Retrieved 08/02/2021 from https://www.accessdata.fda.gov/cdrh_docs/reviews/K200009.pdf
- Friend, B. D., Bailey-Olson, M., Melton, A., Shimano, K. A., Kharbanda, S., Higham, C., Winestone, L. E., Huang, J., Stieglitz, E., & Dvorak, C. C. (2020). The impact of total body irradiation-based regimens on outcomes in children and young adults with acute lymphoblastic leukemia undergoing allogeneic hematopoietic stem cell transplantation. Pediatr Blood Cancer, 67(2), e28079. https://doi.org/10.1002/pbc.28079
- Goicoechea, I., Puig, N., Cedena, M. T., Burgos, L., Cordon, L., Vidriales, M. B., Flores-Montero, J., Gutierrez, N. C., Calasanz, M. J., Ramos, M. M., Lara-Astiaso, D., Vilas-Zornoza, A., Alignani, D., Rodriguez, I., Sarvide, S., Alameda, D., Garces, J. J., Rodriguez, S., Fresquet, V., . . . Paiva, B. (2021). Deep MRD profiling defines outcome and unveils different modes of treatment resistance in standard- and high-risk myeloma. Blood, 137(1), 49-60. https://doi.org/10.1182/blood.2020006731
- Gunning, A., Kumar, S., Williams, C. K., Berger, B. M., Naber, S. P., Gupta, P. B., Del Vecchio Fitz, C., & Kuperwasser, C. (2023). Analytical Validation of NavDx, a cfDNA-Based Fragmentomic Profiling Assay for HPV-Driven Cancers. Diagnostics, 13(4), 725. https://www.mdpi.com/2075-4418/13/4/725
- Hallek, M., Cheson, B. D., Catovsky, D., Caligaris-Cappio, F., Dighiero, G., Dohner, H., Hillmen, P., Keating, M., Montserrat, E., Chiorazzi, N., Stilgenbauer, S., Rai, K. R., Byrd, J. C., Eichhorst, B., O'Brien, S., Robak, T., Seymour, J. F., & Kipps, T. J. (2018). iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood, 131(25), 2745-2760. https://doi.org/10.1182/blood-2017-09-806398
- Hay, K. A., Gauthier, J., Hirayama, A. V., Voutsinas, J. M., Wu, Q., Li, D., Gooley, T. A., Cherian, S., Chen, X., Pender, B. S., Hawkins, R. M., Vakil, A., Steinmetz, R. N., Schoch, G., Chapuis, A. G., Till, B. G., Kiem, H. P., Ramos, J. D., Shadman, M., . . . Turtle, C. J. (2019). Factors associated with durable EFS in adult B-cell ALL patients achieving MRD-negative CR after CD19 CAR T-cell therapy. Blood, 133(15), 1652-1663. https://doi.org/10.1182/blood-2018-11-883710
- Herrera, A. F., Kim, H. T., Kong, K. A., Faham, M., Sun, H., Sohani, A. R., Alyea, E. P., Carlton, V. E., Chen, Y. B., Cutler, C. S., Ho, V. T., Koreth, J., Kotwaliwale, C., Nikiforow, S., Ritz, J., Rodig, S. J., Soiffer, R. J., Antin, J. H., & Armand, P. (2016). Next-generation sequencing-based detection of circulating tumour DNA After allogeneic stem cell transplantation for lymphoma. Br J Haematol, 175(5), 841-850. https://doi.org/10.1111/bjh.14311
- Heuser, M., Ofran, Y., Boissel, N., Brunet Mauri, S., Craddock, C., Janssen, J., Wierzbowska, A., & Buske, C. (2020). Acute myeloid leukaemia in adult patients: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. https://doi.org/10.1016/j.annonc.2020.02.018
- Hoelzer, D., Bassan, R., Dombret, H., Fielding, A., Ribera, J. M., & Buske, C. (2016). Acute lymphoblastic leukaemia in adult patients: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol, 27(suppl 5), v69-v82. https://doi.org/10.1093/annonc/mdw025
- Hofste, L. S. M., Geerlings, M. J., Kamping, E. J., Kouwenhoven, N. D. H., von Rhein, D., Jansen, E. A. M., Garms, L. M., Nagtegaal, I. D., van der Post, R. S., de Wilt, J. H. W., Klarenbeek, B. R., & Ligtenberg, M. J. L. (2023). Clinical Validity of Tumor-Informed Circulating Tumor DNA Analysis in Patients Undergoing Surgery of Colorectal Metastases. Dis Colon Rectum, 66(6), 796-804. https://doi.org/10.1097/DCR.0000000000002443
- Horton, T. M., & Steuber, C. P. (2023, December 19). Risk group stratification and prognosis for acute lymphoblastic leukemia/lymphoblastic lymphoma in children and adolescents. https://www.uptodate.com/contents/risk-group-stratification-and-prognosis-for-acute-lymphoblastic-leukemia-lymphoblastic-lymphoma-in-children-and-adolescents
- Jacome, A. A., & Johnson, B. (2023). Minimal Residual Disease in Colorectal Cancer: Are We Finding the Needle in a Haystack? Cells, 12(7). https://doi.org/10.3390/cells12071068
- Kater, A. P., Seymour, J. F., Hillmen, P., Eichhorst, B., Langerak, A. W., Owen, C., Verdugo, M., Wu, J., Punnoose, E. A., Jiang, Y., Wang, J., Boyer, M., Humphrey, K., Mobasher, M., & Kipps, T. J. (2019). Fixed Duration of Venetoclax-Rituximab in Relapsed/Refractory Chronic Lymphocytic Leukemia Eradicates Minimal Residual Disease and Prolongs Survival: Post-Treatment Follow-Up of the MURANO Phase III Study. J Clin Oncol, 37(4), 269-277. https://doi.org/10.1200/jco.18.01580
- Khaddour, K., Zhou, A., Butt, O. H., Budde, G., Malashevich, A. K., & Ansstas, G. (2022). Case report: Real-world experience using a personalized cancer-specific circulating tumor DNA assay in different metastatic melanoma scenarios. Front Oncol, 12, 978996. https://doi.org/10.3389/fonc.2022.978996
- Kim, H. Y., Yoo, I. Y., Lim, D. J., Kim, H. J., Kim, S. H., Yoon, S. E., Kim, S. J., Cho, D., & Kim, K. (2022). Clinical Utility of Next-Generation Flow-Based Minimal Residual Disease Assessment in Patients with Multiple Myeloma. Ann Lab Med, 42(5), 558-565. https://doi.org/10.3343/alm.2022.42.5.558
- Kumar, S., Paiva, B., Anderson, K. C., Durie, B., Landgren, O., Moreau, P., Munshi, N., Lonial, S., Blade, J., Mateos, M. V., Dimopoulos, M., Kastritis, E., Boccadoro, M., Orlowski, R., Goldschmidt, H., Spencer, A., Hou, J., Chng, W. J., Usmani, S. Z., . . . Avet-Loiseau, H. (2016). International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol, 17(8), e328-e346. https://doi.org/10.1016/s1470-2045(16)30206-6
- Lee, S., Kim, D. W., Cho, B., Kim, Y. J., Kim, Y. L., Hwang, J. Y., Park, Y. H., Shin, H. J., Park, C. Y., Min, W. S., Kim, H. K., & Kim, C. C. (2003). Risk factors for adults with Philadelphia-chromosome-positive acute lymphoblastic leukaemia in remission treated with allogeneic bone marrow transplantation: the potential of real-time quantitative reverse-transcription polymerase chain reaction. Br J Haematol, 120(1), 145-153. https://doi.org/10.1046/j.1365-2141.2003.03988.x
- Loupakis, F., Sharma, S., Derouazi, M., Murgioni, S., Biason, P., Rizzato, M. D., Rasola, C., Renner, D., Shchegrova, S., Koyen Malashevich, A., Malhotra, M., Sethi, H., Zimmermann, B. G., Aleshin, A., Moshkevich, S., Billings, P. R., Sedgwick, J. D., Schirripa, M., Munari, G., . . . Fassan, M. (2021). Detection of Molecular Residual Disease Using Personalized Circulating Tumor DNA Assay in Patients With Colorectal Cancer Undergoing Resection of Metastases. JCO Precis Oncol, 5. https://doi.org/10.1200/PO.21.00101
- Luskin, M. R., Murakami, M. A., Manalis, S. R., & Weinstock, D. M. (2018). Targeting minimal residual disease: a path to cure? Nat Rev Cancer, 18(4), 255-263. https://doi.org/10.1038/nrc.2017.125
- Madzo, J., Zuna, J., Muzíková, K., Kalinová, M., Krejcí, O., Hrusák, O., Otová, B., Starý, J., & Trka, J. (2003). Slower molecular response to treatment predicts poor outcome in patients
- with TEL/AML1 positive acute lymphoblastic leukemia: prospective real-time quantitative reverse transcriptase-polymerase chain reaction study. Cancer, 97(1), 105-113. https://doi.org/10.1002/cncr.11043
- Medina, A., Puig, N., Flores-Montero, J., Jimenez, C., Sarasquete, M. E., Garcia-Alvarez, M., Prieto-Conde, I., Chillon, C., Alcoceba, M., Gutierrez, N. C., Oriol, A., Rosinol, L., Blade, J., Gironella, M., Hernandez, M. T., Gonzalez-Calle, V., Cedena, M. T., Paiva, B., San-Miguel, J. F., . . . Garcia-Sanz, R. (2020). Comparison of next-generation sequencing (NGS) and next-generation flow (NGF) for minimal residual disease (MRD) assessment in multiple myeloma. Blood Cancer J, 10(10), 108. https://doi.org/10.1038/s41408-020-00377-0
- Mikhael, J., Ismaila, N., Cheung, M. C., Costello, C., Dhodapkar, M. V., Kumar, S., Lacy, M., Lipe, B., Little, R. F., Nikonova, A., Omel, J., Peswani, N., Prica, A., Raje, N., Seth, R., Vesole, D. H., Walker, I., Whitley, A., Wildes, T. M., . . . Martin, T. (2019). Treatment of Multiple Myeloma: ASCO and CCO Joint Clinical Practice Guideline. Journal of Clinical Oncology, 37(14), 1228-1263. https://doi.org/10.1200/JCO.18.02096
- Moding, E. J., Nabet, B. Y., Alizadeh, A. A., & Diehn, M. (2021). Detecting Liquid Remnants of Solid Tumors: Circulating Tumor DNA Minimal Residual Disease. Cancer Discov, 11(12), 2968-2986. https://doi.org/10.1158/2159-8290.CD-21-0634
- Natera. (2024). Signatera™ Transforming the management of cancer with personalized testing. https://www.natera.com/oncology/signatera-advanced-cancer-detection/
- NavDx. (2024). Get Answers to Patients' Frequently Asked Questions (FAQs) Regarding NavDx®. https://navdx.com/patient-faqs/
- NCCN. (2024a, July 19, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Acute Lymphoblastic Leukemia Version 2.2024. https://www.nccn.org/professionals/physician_gls/pdf/all.pdf
- NCCN. (2024b, May 17, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Acute Myeloid Leukemia Version 3.2024. https://www.nccn.org/professionals/physician_gls/pdf/aml.pdf
- NCCN. (2024c, October 1, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Chronic Lymphocytic Leukemia / Small Lymphocytic Leukemia Version 1.2025. https://www.nccn.org/professionals/physician_gls/pdf/cll.pdf
- NCCN. (2024d, September 26, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Hairy Cell Leukemia Version 1.2025. https://www.nccn.org/professionals/physician_gls/pdf/hairy_cell.pdf
- NCCN. (2024e, September 17, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Multiple Myeloma Version 1.2025. https://www.nccn.org/professionals/physician_gls/pdf/myeloma.pdf
- NCCN. (2024f, August 28, 2024). NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines) Pediatric Acute Lymphoblastic Leukemia Version 1.2025. https://www.nccn.org/professionals/physician_gls/pdf/ped_all.pdf
- Nguyen Hoang, T. P., Nguyen, T. A., Tran, N. H. B., Nguyen Hoang, V. A., Thi Dao, H. T., Tran, V. U., Nguyen, Y. N., Nguyen, A. T., Nguyen Thi, C. T., Do Thi, T. T., Nguyen, D. S., Nguyen, H. N., Giang, H., & Tu, L. N. (2024). Analytical validation and clinical utilization of K-4CARE™: a comprehensive genomic profiling assay with personalized MRD detection. Front Mol Biosci, 11, 1334808. https://doi.org/10.3389/fmolb.2024.1334808
- O'Sullivan, H. M., Feber, A., & Popat, S. (2023). Minimal Residual Disease Monitoring in Radically Treated Non-Small Cell Lung Cancer: Challenges and Future Directions. Onco Targets Ther, 16, 249-259. https://doi.org/10.2147/OTT.S322242
- Perrot, A., Lauwers-Cances, V., Corre, J., Robillard, N., Hulin, C., Chretien, M. L., Dejoie, T., Maheo, S., Stoppa, A. M., Pegourie, B., Karlin, L., Garderet, L., Arnulf, B., Doyen, C., Meuleman, N., Royer, B., Eveillard, J. R., Benboubker, L., Dib, M., . . . Munshi, N. (2018). Minimal residual disease negativity using deep sequencing is a major prognostic factor in multiple myeloma. Blood, 132(23), 2456-2464. https://doi.org/10.1182/blood-2018-06-858613
- Rai, K. R., & Stilgenbauer, S. (2024, February 13). Evaluating response to treatment of chronic lymphocytic leukemia. https://www.uptodate.com/contents/evaluating-response-to-treatment-of-chronic-lymphocytic-leukemia
- Rajkumar, S. V. (2023, September 25, 2023). Multiple myeloma: Evaluating response to treatment. https://www.uptodate.com/contents/multiple-myeloma-evaluating-response-to-treatment
- Rawstron, A. C., Fazi, C., Agathangelidis, A., Villamor, N., Letestu, R., Nomdedeu, J., Palacio, C., Stehlikova, O., Kreuzer, K. A., Liptrot, S., O'Brien, D., de Tute, R. M., Marinov, I., Hauwel, M., Spacek, M., Dobber, J., Kater, A. P., Gambell, P., Soosapilla, A., . . . Ghia, P. (2016). A complementary role of multiparameter flow cytometry and high-throughput sequencing for minimal residual disease detection in chronic lymphocytic leukemia: an European Research Initiative on CLL study. Leukemia, 30(4), 929-936. https://doi.org/10.1038/leu.2015.313
- Reinert, T., Henriksen, T. V., Christensen, E., Sharma, S., Salari, R., Sethi, H., Knudsen, M., Nordentoft, I., Wu, H. T., Tin, A. S., Heilskov Rasmussen, M., Vang, S., Shchegrova, S., Frydendahl Boll Johansen, A., Srinivasan, R., Assaf, Z., Balcioglu, M., Olson, A., Dashner, S., . . . Lindbjerg Andersen, C. (2019). Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol, 5(8), 1124-1131. https://doi.org/10.1001/jamaoncol.2019.0528
- Robak, T., Matutes, E., Catovsky, D., Zinzani, P. L., & Buske, C. (2015). Hairy cell leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol, 26 Suppl 5, v100-107. https://doi.org/10.1093/annonc/mdv200
- Royston, D. J., Gao, Q., Nguyen, N., Maslak, P., Dogan, A., & Roshal, M. (2016). Single-Tube 10-Fluorochrome Analysis for Efficient Flow Cytometric Evaluation of Minimal Residual Disease in Plasma Cell Myeloma. Am J Clin Pathol, 146(1), 41-49. https://doi.org/10.1093/ajcp/aqw052
- Sato, S., Nakamura, Y., Oki, E., & Yoshino, T. (2023). Molecular Residual Disease-guided Adjuvant Treatment in Resected Colorectal Cancer: Focus on CIRCULATE-Japan. Clin Colorectal Cancer, 22(1), 53-58. https://doi.org/10.1016/j.clcc.2022.12.001
- Schuurhuis, G. J., Heuser, M., Freeman, S., Bene, M. C., Buccisano, F., Cloos, J., Grimwade, D., Haferlach, T., Hills, R. K., Hourigan, C. S., Jorgensen, J. L., Kern, W., Lacombe, F., Maurillo, L., Preudhomme, C., van der Reijden, B. A., Thiede, C., Venditti, A., Vyas, P., . . . Ossenkoppele, G. J. (2018). Minimal/measurable residual disease in AML: a consensus document from the European LeukemiaNet MRD Working Party. Blood, 131(12), 1275-1291. https://doi.org/10.1182/blood-2017-09-801498
- Stock, W., & Estrov, Z. (2022a, June 16, 2022). Clinical use of measurable residual disease detection in acute lymphoblastic leukemia. https://www.uptodate.com/contents/clinical-use-of-measurable-residual-disease-detection-in-acute-lymphoblastic-leukemia
- Stock, W., & Estrov, Z. (2022b, March 7, 2022). Detection of measurable residual disease in acute lymphoblastic leukemia. https://www.uptodate.com/contents/detection-of-measurable-residual-disease-in-acute-lymphoblastic-leukemia
- Subhash, V. V., Huang, L., Kamili, A., Wong, M., Chen, D., Venn, N. C., Atkinson, C., Mayoh, C., Venkat, P., Tyrrell, V., Marshall, G. M., Cowley, M. J., Ekert, P. G., Norris, M. D., Haber, M., Henderson, M. J., Sutton, R., Fletcher, J. I., & Trahair, T. N. (2022). Whole-genome sequencing facilitates patient-specific quantitative PCR-based minimal residual disease monitoring in acute lymphoblastic leukaemia, neuroblastoma and Ewing sarcoma. Br J Cancer, 126(3), 482-491. https://doi.org/10.1038/s41416-021-01538-z
- Sullivan, B. G., Dayyani, F., & Senthil, M. (2023). ASO Author Reflections: Challenges of Circulating Tumor DNA in the Management of Gastrointestinal Peritoneal Carcinomatosis. Ann Surg Oncol, 30(1), 285-286. https://doi.org/10.1245/s10434-022-12543-8
- Theunissen, P., Mejstrikova, E., Sedek, L., van der Sluijs-Gelling, A. J., Gaipa, G., Bartels, M., Sobral da Costa, E., Kotrová, M., Novakova, M., Sonneveld, E., Buracchi, C., Bonaccorso, P., Oliveira, E., Te Marvelde, J. G., Szczepanski, T., Lhermitte, L., Hrusak, O., Lecrevisse, Q., Grigore, G. E., . . . van der Velden, V. H. (2017). Standardized flow cytometry for highly sensitive MRD measurements in B-cell acute lymphoblastic leukemia. Blood, 129(3), 347-357. https://doi.org/10.1182/blood-2016-07-726307
- Thompson, P. A., Srivastava, J., Peterson, C., Strati, P., Jorgensen, J. L., Hether, T., Keating, M. J., O'Brien, S. M., Ferrajoli, A., Burger, J. A., Estrov, Z., Jain, N., & Wierda, W. G. (2019). Minimal residual disease undetectable by next-generation sequencing predicts improved outcome in CLL after chemoimmunotherapy. Blood, 134(22), 1951-1959. https://doi.org/10.1182/blood.2019001077
- Thörn, I., Forestier, E., Botling, J., Thuresson, B., Wasslavik, C., Björklund, E., Li, A., Lindström-Eriksson, E., Malec, M., Grönlund, E., Torikka, K., Heldrup, J., Abrahamsson, J., Behrendtz, M., Söderhäll, S., Jacobsson, S., Olofsson, T., Porwit, A., Lönnerholm, G., . . . Sundström, C. (2011). Minimal residual disease assessment in childhood acute lymphoblastic leukaemia: a Swedish multi-centre study comparing real-time polymerase chain reaction and multicolour flow cytometry. Br J Haematol, 152(6), 743-753. https://doi.org/10.1111/j.1365-2141.2010.08456.x
- Tie, J., Cohen, J. D., Wang, Y., Christie, M., Simons, K., Lee, M., Wong, R., Kosmider, S., Ananda, S., McKendrick, J., Lee, B., Cho, J. H., Faragher, I., Jones, I. T., Ptak, J., Schaeffer, M. J., Silliman, N., Dobbyn, L., Li, L., . . . Gibbs, P. (2019). Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol, 5(12), 1710-1717. https://doi.org/10.1001/jamaoncol.2019.3616
- Tie, J., Wang, Y., Tomasetti, C., Li, L., Springer, S., Kinde, I., Silliman, N., Tacey, M., Wong, H. L., Christie, M., Kosmider, S., Skinner, I., Wong, R., Steel, M., Tran, B., Desai, J., Jones, I., Haydon, A., Hayes, T., . . . Gibbs, P. (2016). Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med, 8(346), 346ra392. https://doi.org/10.1126/scitranslmed.aaf6219
- van der Velden, V. H. J., Hochhaus, A., Cazzaniga, G., Szczepanski, T., Gabert, J., & van Dongen, J. J. M. (2003). Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects. Leukemia, 17(6), 1013-1034. https://doi.org/10.1038/sj.leu.2402922
- Wang, Y., Li, L., Cohen, J. D., Kinde, I., Ptak, J., Popoli, M., Schaefer, J., Silliman, N., Dobbyn, L., Tie, J., Gibbs, P., Tomasetti, C., Kinzler, K. W., Papadopoulos, N., Vogelstein, B., & Olsson, L. (2019). Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol, 5(8), 1118-1123. https://doi.org/10.1001/jamaoncol.2019.0512
- Wang, Z., Guo, M., Zhang, Y., Xu, S., Cheng, H., Wu, J., Zhang, W., Hu, X., Yang, J., Wang, J., & Tang, G. (2019). The applicability of multiparameter flow cytometry for the detection of
- minimal residual disease using different-from-normal panels to predict relapse in patients with acute myeloid leukemia after allogeneic transplantation. Int J Lab Hematol, 41(5), 607-614. https://doi.org/10.1111/ijlh.13070
- Wood, B., Wu, D., Crossley, B., Dai, Y., Williamson, D., Gawad, C., Borowitz, M. J., Devidas, M., Maloney, K. W., Larsen, E., Winick, N., Raetz, E., Carroll, W. L., Hunger, S. P., Loh, M. L., Robins, H., & Kirsch, I. (2018). Measurable residual disease detection by high-throughput sequencing improves risk stratification for pediatric B-ALL. Blood, 131(12), 1350-1359. https://doi.org/10.1182/blood-2017-09-806521
- Zhang, S., Brazel, D., Kumar, P., Schafer, L. N., Eidenschink, B., Senthil, M., & Dayyani, F. (2021). Utility of tumor-informed circulating tumor DNA in the clinical management of gastrointestinal malignancies. J Gastrointest Oncol, 12(6), 2643-2652. https://doi.org/10.21037/jgo-21-484
- Zhong, R., Gao, R., Fu, W., Li, C., Huo, Z., Gao, Y., Lu, Y., Li, F., Ge, F., Tu, H., You, Z., He, J., & Liang, W. (2023). Accuracy of minimal residual disease detection by circulating tumor DNA profiling in lung cancer: a meta-analysis. BMC Med, 21(1), 180. https://doi.org/10.1186/s12916-023-02849-z
Coding Section
| Code |
Number |
Description |
| CPT |
88184 |
Flow cytometry, cell surface, cytoplasmic, or nuclear marker, technical component only; first marker |
|
|
88185 |
Flow cytometry, cell surface, cytoplasmic, or nuclear marker, technical component only; each additional marker (List separately in addition to code for first marker) |
|
|
81479 |
Unlisted molecular pathology procedure |
|
|
0171U |
Targeted genomic sequence analysis panel, acute myeloid leukemia, myelodysplastic syndrome, and myeloproliferative neoplasms, DNA analysis, 23 genes, interrogation for sequence variants, rearrangements and minimal residual disease, reported as presence/absence |
| 0306U | Oncology (minimal residual disease [MRD]), next-generation targeted sequencing analysis, cell-free DNA, initial (baseline) assessment to determine a patient-specific panel for future comparisons to evaluate for MRD Proprietary test: Invitae PCM Tissue Profiling and MRD Baseline Assay Lab/Manufacturer: Invitae Corporation |
|
| 0307U | Oncology (minimal residual disease [MRD]), next-generation targeted sequencing analysis of a patient-specific panel, cell-free DNA, subsequent assessment with comparison to previously analyzed patient specimens to evaluate for MRD Proprietary test: Invitae PCM MRD Monitoring Lab/Manufacturer: Invitae Corporation |
|
| 0340U | Oncology (pan-cancer), analysis of minimal residual disease (MRD) from plasma, with assays personalized to each patient based on prior next-generation sequencing of the patient's tumor and germline DNA, reported as absence or presence of MRD, with disease-burden correlation, if appropriate Protietary test: Signatera™ Lab/Manufacturer: Natera Inc. |
|
| 0356U | Oncology (oropharyngeal or anal), evaluation of 17 DNA biomarkers using droplet digital PCR (ddPCR), cell-free DNA, algorithm reported as a prognostic risk score for cancer recurrence Proprietary test: NavDx® Lab/Manufacturer: Naveris Inc. |
|
| 0364U (effective 04/01/2023) | Oncology (hematolymphoid neoplasm), genomic sequence analysis using multiplex (PCR) and next-generation sequencing with algorithm, quantification of dominant clonal sequence(s), reported as presence or absence of minimal residual disease (MRD) with quantitation of disease burden, when appropriate Proprietary test: clonoSEQ® Assay |
|
| 0422U (effective 01/01/2024) | Oncology (pan-solid tumor), analysis of DNA biomarker response to anti-cancer therapy using cell-free circulating DNA, biomarker comparison to a previous baseline pre-treatment cell-free circulating DNA analysis using next-generation sequencing, algorithm reported as a quantitative change from baseline, including specific alterations, if appropriate. |
|
| 0467U (effective 07/01/2024) | Oncology (bladder), DNA, next generation sequencing (NGS) of 60 genes and whole genome aneuploidy, urine, algorithms reported as minimal residual disease (MRD) status positive or negative and quantitative disease burden Protietary test: UroAmp MRD Lab/Manufacturer: Convergent Genomics Inc., Convergent Genomics Inc. |
|
| 0470U (effective 07/01/2024) | Oncology (oropharyngeal), detection of minimal residual disease by next-generation sequencing (NGS) based quantitative evaluation of 8 DNA targets, cell-free HPV 16 and 18 DNA from plasma Protietary test: HPV-SEQ Test Lab/Manufacturer: Sysmex Inostics Inc. |
|
| ICD10 Diagnosis Code |
C81.00 – C81.99 |
Hodgkin lymphoma |
|
|
C82.00 – C82.99 |
Follicular lymphoma |
|
|
C83.00 – C83.99 |
Non-follicular lymphoma |
|
|
C84.00 – C84.99 |
Mature T/NK-cell lymphomas |
|
|
C85.10 – C85.99 |
Other specified and unspecified types of non-Hodgkin lymphoma |
|
|
C86.0 – C86.6 |
Other specified types of T/NK-cell lymphoma |
|
|
C88.0 – C88.9 |
Malignant immunoproliferative diseases and certain other B-cell lymphomas |
|
|
C90.00 – C90.32 |
Multiple myeloma and malignant plasma cell neoplasms |
|
|
C91.00 – C91.92 |
Lymphoid leukemia |
|
|
C92.00 – C92.92 |
Myeloid leukemia |
|
|
C93.00 – C93.92 |
Monocyte leukemia |
|
|
C94.00 – C94.82 |
Other leukemias of specified cell type |
|
|
C95.00 – C95.92 |
Leukemia of unspecified cell type |
|
|
C96.0 – C96.9 |
Other and unspecified malignant neoplasms of lymphoid, hematopoietic and related tissue |
|
|
Z85.79
|
Personal history of other malignant neoplasms of lymphoid, hematopoietic and related tissues |
Procedure and diagnosis codes on Medical Policy documents are included only as a general reference tool for each policy. They may not be all-inclusive.
This medical policy was developed through consideration of peer-reviewed medical literature generally recognized by the relevant medical community, U.S. FDA approval status, nationally accepted standards of medical practice and accepted standards of medical practice in this community and other nonaffiliated technology evaluation centers, reference to federal regulations, other plan medical policies, and accredited national guidelines.
"Current Procedural Terminology © American Medical Association. All Rights Reserved"
History From 2020 Forward
| 01/22/2025 | Interim review, adding new criteria #3 and updating new criteria #4. Also updating table of term, rationale, and references. Add CPT 0356U |
| 08/01/2024 | Annual review, no change to policy intent. Updating description, table of Terminology, rationale, references, and coding. |
| 06/25/2024 | Interim review to add PLA codes 0467U and 0470U to coding section. |
| 01/11/2024 | Added code 0422U effective 01/01/2024 |
| 11/03/2023 | Annual review, entire policy updated for clarity and consistency. Policy criteria compressed into 3 statements |
| 03/08/2023 | Adding code ‘0364U’, effective date 04012023 |
| 12/20/2022 | Annual review, no change to policy. Maintaining as written. |
| 10/01/2021 |
Annual review, no change to policy intent. Updating background, rationale, references and coding |
| 08/09/2021 |
Updating Annual review date to coincide with Avalon. No other changes. |
| 01/01/2021 |
New Policy |