Prenatal Screening (Genetic) - CAM 358
Description
Prenatal screening encompasses any testing done to determine the health status of the pregnant individual and/or fetus. Genetic prenatal screening encompasses screening to determine the risk of fetal abnormalities, including genetic and developmental abnormalities. Any individual undergoing screening tests, especially genetic carrier screenings, must realize the limitations of screening tests and the difference between screening and diagnostic testing. Screening refers to testing of asymptomatic or healthy individuals to search for a condition that may affect the pregnancy or individual, whereas diagnostic testing is used to either confirm or refute true abnormalities in an individual.1,2
This policy addresses broad prenatal genetic screening, as well as screening for conditions not addressed in condition-specific policies. For situations in which prenatal and pre-conception screening may be discussed in further detail, please see the “Related Policies” section of this policy document.
Terms such as male and female are used when necessary to refer to sex assigned at birth.
Regulatory Status
The FDA has approved many tests for conditions that can be included in prenatal screening, such as HSV, chlamydia, gonorrhea, syphilis, and diabetes. Additionally, many labs have developed specific tests that they must validate and perform in-house. These laboratory-developed tests (LDTs) are regulated by the Centers for Medicare and Medicaid (CMS) as high-complexity tests under the Clinical Laboratory Improvement Amendments of 1988 (CLIA ’88). LDTs are not approved or cleared by the U. S. Food and Drug Administration; however, FDA clearance or approval is not currently required for clinical use.
Policy
Application of coverage criteria is dependent upon an individual’s benefit coverage at the time of the request.
- For individuals who are pregnant or who are capable of becoming pregnant and seeking pre-conception care, single gene or multi-gene panel screening of the individual for conditions classified through ACMG as a Tier 1, Tier 2, or Tier 3 condition (see Note 1) is considered MEDCIALLY NECESSARY.
- For pregnant individuals and those capable of becoming pregnant who come from a family with a genetic disorder for which a properly validated test is available, the following testing is considered MEDCIALLY NECESSARY:
- Testing restricted to the known mutation.
- Comprehensive genetic testing, including multi-gene panel testing specific to the familial genetic disorder, when the specific familial mutation is unknown.
- For individuals planning a pregnancy with a reproductive partner who is known or found to be a carrier of a recessively inherited disorder, genetic testing specific to the genes for which the reproductive partner is a carrier is considered MEDCIALLY NECESSARY.
- For RHD negative pregnant individuals, fetal RHD genotyping using maternal plasma is considered MEDCIALLY NECESSARY.
- For fetuses with a high risk for a genetic disorder, prenatal genetic testing using cells obtained for diagnostic cytogenetic testing (i.e., amniocentesis or chorionic villus sampling [CVS]) is considered MEDCIALLY NECESSARY.
- Carrier screening for the same gene more than once per lifetime is considered NOT MEDCIALLY NECESSARY.
The following does not meet coverage criteria due to a lack of available published scientific literature confirming that the test(s) is/are required and beneficial for the diagnosis and treatment of an individual’s illness.
- To screen for single-gene mutations (i.e., autosomal recessive, autosomal dominant, X-linked) in the fetus, the use of non-invasive prenatal screening (NIPS) is considered NOT MEDCIALLY NECESSARY.
- For all other inherited medical disorders not meeting the above criteria, pre-conceptional or prenatal genetic testing is consideredNOT MEDCIALLY NECESSARY.
NOTES:
Note 1: Please see the “Guidelines and Recommendations” section of this policy for ACMG’s tiered system based on carrier frequency (Tables 1-6).
Note 2: For two or more gene tests being run on the same platform, please refer to CAM 235-Reimbursement Policy.
Table of Terminology
| Term |
Definition |
| ACMG |
American College of Medical Genetics and Genomics |
| ACOG |
American College of Obstetricians and Gynecologists |
| ADA |
American Diabetes Association |
| BSH |
British Society for Haematology |
| CAP |
College of American Pathologists |
| CAVD |
Congenital absence of the vas deferens |
| cfDNA |
Cell-free deoxyribonucleic acid |
| CFTR |
Cystic Fibrosis transmembrane conductance regulator |
| CMA |
Chromosomal microarray |
| CNVs |
Copy number variants |
| CVS |
Chorionic villus sampling |
| DMD |
Dystrophin |
| DNA |
Deoxyribonucleic acid |
| dSGD |
Dominant single-gene disorder |
| FRAXE |
Fragile site, folic acid type, rare, Fra(X)(Q28) E |
| GJB6 |
Gap junction protein |
| HBB |
Human beta-globin gene |
| HDFN |
Hemolytic disease of the fetus and newborn |
| ISPD |
International Society for Prenatal Diagnosis |
| MMS |
Microdeletion/microduplication syndromes |
| NGS |
Next-generation sequencing |
| NIPS-Plus |
Expanded non-invasive prenatal screening |
| NIPT |
Non-invasive prenatal testing |
| NT |
Nuchal translucency |
| PPVs |
Positive predictive values |
| PQF |
Perinatal Quality Foundation |
| RBC |
Red blood cells |
| RHD |
Rh blood group D antigen |
| Rhlg |
Human Rho(D) immune globulin |
| sgNIPS |
Single-gene non-invasive prenatal screening |
| SMA |
Spinal muscular atrophy |
| SMFM |
Society for Maternal Fetal Medicine |
| SMN1 |
Survival of motor neuron 1 |
| SNP |
Single nucleotide polymorphism |
| TMRC |
Transfusion Medicine Resource Committee |
Rationale
Prenatal screening is a part of overall prenatal care to promote optimal care of both mother and baby. Prenatal screening allows for assessment and monitoring of the fetus for the presence of congenital defects or disease. Various professional medical organizations provide guidelines for prenatal screening. “Screening is an offer on the initiative of the health system or society, rather than a medical intervention in answer to a patient’s complaint or health problem. Screening aims at obtaining population health gains through early detection that enables prevention or treatment.”3
Genetic screening tests, including carrier screening for genetic mutations and fetal testing for chromosomal aneuploidy, can be a part of prenatal screening. Aneuploidy screening may be performed on cell-free DNA in maternal circulation or by examining maternal serum levels of specific biochemical markers for trisomy.2 These non-invasive prenatal testing (NIPT) can decrease the number of more invasive procedures and the risks of unwanted side effects. A chromosomal microarray (CMA) can screen all chromosomes in a single test and “can detect many very small variants that cannot be detected by traditional karyotyping.”3 The American College of Obstetricians and Gynecologists (ACOG) recommends CMA for instances where the ultrasound of a fetus shows a major structural abnormality.4 CMA in this situation should be performed on DNA from amniotic fluid, chorionic villus cells, or cord blood, rather than on maternal serum cell-free DNA since the process does not include an amplification step and the maternal DNA signal would be many times higher than the fetal DNA.5
Several companies, such as LabCorp, have developed panels to test for potential genetic mutations in pregnant individuals, or in individuals planning to become pregnant. This includes the Inheritest® Carrier Screening which encompasses six different panels to identify potential genetic mutations. These six panels include the Inheritest® 500 PLUS Panel (which screens 525 genes for several clinically relevant genetic disorders), the Inheritest® Comprehensive Panel (which screens for more than 110 disorders), the Inheritest® Ashkenazi Jewish Panel (which screens for more than 40 Ashkenazi Jewish related disorders), the Inheritest® Society-Guided Panel (which screens for more than 13 disorders highlighted in the American College of Medical Genetics and Genomics and the American Congress of Obstetricians and Gynecologists guidelines), the Inheritest® Core Panel (which screens for cystic fibrosis, fragile X syndrome, and spinal muscular atrophy), and the Inheritest® CF/SMA (spinal muscular atrophy) Panel (which screens only for cystic fibrosis and spinal muscular atrophy).6
Additionally, the company BillionToOne has created a NIPS test. UNITY Complete® uses cell-free DNA from a maternal blood draw and assesses for seven aneuploidies (trisomy 21, trisomy 18, trisomy 13, monosomy X, XXX, XXY, and XYY), and five recessive conditions (cystic fibrosis, spinal muscular atrophy, sickle cell disease, alpha thalassemia, and beta-thalassemia). This screen functions in a sequential manner. First, the screen uses NGS of genomic DNA to assess maternal carrier status for genes associated with the most common single-gene recessive disorders. If the pregnant individual is identified as a carrier for a pathogenic variant in one or more of these genes, the sample is then reflexed to single-gene non-invasive prenatal screening (sgNIPS). In sgNIPS, NGS is performed on cfDNA extracted from the original blood sample, from which fetal risk is calculated. Fetal risk assessment is summarized as low-risk (fetal risk 1/500), high risk (fetal risk >1/4), increased risk or decreased risk (fetal risk between 1/500 and 1/4), or no result.7,8
Red blood cell antigen discrepancy between a mother and fetus may also occur during pregnancy. This is known as hemolytic disease of the fetus and newborn (HDFN), and causes maternal antibodies to destroy the red blood cells of the neonate or fetus.9 Alloimmunization is the immune response which occurs in the mother due to foreign antigens after exposure to genetically foreign cells, occurring almost exclusively in mothers with type O blood. However, while ABO blood type incompatibility is identified in almost 15% of pregnancies, HDFN is only identified in approximately 4% of pregnancies.9 Another important inherited antigen sometimes found on the surface of red blood cells is known as the Rhesus (Rh)D antigen. During pregnancy and delivery, individuals who are RhD-negative may be exposed to RhD-positive fetal cells, which can lead to the development of anti-RhD antibodies. This exposure typically happens during delivery and affects subsequent pregnancies; infants with RhD incompatibility tend to experience a more severe form of HDFN than those with ABO incompatibility. The clinical presentation of HDFN may be mild (such as hyperbilirubinemia with mild to moderate anemia) to severe and life-threatening anemia (such as hydrops fetalis). Less severely affected infants may develop hyperbilirubinemia within the first day of life; infants with RhD HDFN may also present with symptomatic anemia requiring a blood transfusion. In more severe cases, infants with severe life-threatening anemia, such as hydrops fetalis, may exhibit shock at delivery requiring an emergent blood transfusion.9
The administration of anti-D immune globulin has been able to dramatically reduce, but not eliminate, the number of RhD alloimmunization cases. “Anti-D immune globulin is manufactured from pooled plasma selected for high titers of IgG antibodies to D-positive erythrocytes.”10 Before the development of this anti-D immune globulin, it has been reported that 16% of pregnant RhD-negative individuals with two deliveries of RhD-positive ABO-compatible infants became alloimmunized. However, this rate falls to 1-2% with routine postpartum administration of a single dose of anti-D immune globulin. An additional administration in the third trimester of pregnancy further reduces the incidents of alloimmunization to 0.1-0.3%.10
Fetal RhD genotyping using cell-free fetal DNA from maternal plasma can be performed to identify fetal blood type most accurately after 11 weeks of gestation. While the United States has not implemented fetal RhD genotyping for routine prophylaxis and fetal monitoring protocols, several European countries, such as Denmark, the Netherlands, England, Sweden, France and Finland, do utilize fetal RhD determination so that the administration of anti-D immune globulin can be avoided when an RhD-negative fetus is identified.10 Daniels, et al. (2007) report that approximately 40% of RhD-negative pregnant individuals are carrying a RhD-negative fetus; genotypic screening would, therefore, be very valuable in preventing these individuals from receiving unnecessary anti-D immune globulin. Kent, et al. (2014) suggest that the administration of anti-D immune globulin to one third of pregnant individuals who do not require this administration is unethical, and that the availability of RhD genotyping to all RhD-negative pregnant individuals would assist in more informed choices being made regarding anti-D immune globulin administration. Finning, et al. (2008) agree with the previous statements, declaring that “high throughput RHD genotyping of fetuses in all RhD negative [individuals] is feasible and would substantially reduce unnecessary administration of anti-RhD immunoglobulin to RhD negative pregnant [individuals] with an RhD negative fetus.”
Analytical Validity
A prospective cohort study by de Haas, et al. (2016) completed a nationwide program in the Netherlands to determine the sensitivity of fetal RhD screening for the safe guidance of targeted anti-immune globulin prophylaxis. A total of 25,789 RhD-negative pregnant individuals participated in this study. Fetal testing for the RHD gene was assessed in the 27th week of pregnancy. Fetal RHD test results were compared to serological cord blood results after birth. “Sensitivity for detection of fetal RHD was 99.94% (95% confidence interval 99.89% to 99.97%) and specificity was 97.74% (97.43% to 98.02%). Nine false-negative results for fetal RHD testing were registered (0.03%, 95% confidence interval 0.01% to 0.06%).”14 They conclude that fetal RhD testing is a highly reliable testing method.
Manfroi, et al. (2018) completed fetal RhD genotyping with real-time polymerase chain reaction (qPCR) using cell-free fetal DNA extracted from maternal plasma. A commercial multiple-exon assay was used to determine fetal RHD genotypic accuracy. A total of 367 plasma samples obtained between the 24th and 28th weeks of pregnancy were used for this study. Neonatal results were available for 284 of the pregnancies. Sensitivity was reported at 100% and specificity at 97.5%. The diagnostic accuracy was 96.1% with the inclusion of 9/284 inconclusive results.15 The authors conclude that this is therefore an accurate and reliable tool for targeted prenatal immunoprophylaxis.
Clinical Utility and Validity
Education and counseling are a key factor in prenatal screening and diagnostic tests. Yesilcinar and Guvenc (2021) found that a proactive intervention approach decreased anxiety and decisional conflict in the pregnant individual and increased attitudes towards the tests, having a positive effect on the pregnant individual’s knowledge level and decision satisfaction. This allowed the individual to make more informed decisions, such as opting to have screening and diagnostic testing performed. Decreasing anxiety during pregnancy is beneficial to the fetus and individuals receiving educational intervention showed decreased anxiety when receiving genetic screening results as compared to individuals not receiving the same intervention.16 Migliorini, et al. (2020) have also reported that the use of cell-free DNA (cfDNA) screening, combined with a detailed ultrasound examination, as a first-trimester risk assessment is associated with improved maternal reassurance and satisfaction and decreased anxiety, as compared to individuals who received standard first-trimester combined screening with nuchal translucency (NT) and biochemistry.17
Biro, et al. (2020) report on a non-invasive prenatal testing method for congenital heart disease, utilizing the measurement of cell-free nucleic acid and protein biomarkers in maternal blood. Congenital heart disease is considered the most common fetal malformation. While prenatal ultrasonography is currently used to diagnose congenital heart disease, it is not the most accurate method. After a large review completed with PubMed and Web of Sciences databases, the authors conclude that most fetal congenital heart disease related disorders can be diagnosed by NIPT techniques. Further, cell-free RNAs and circulating proteins are potential biomarkers for fetal congenital heart disease and may be able to improve the detection rate in early pregnancies.18
A study by Persico, et al. (2016) investigated the clinical implication of cfDNA testing in high-risk pregnancies. In their cohort of 259 singleton pregnancies, cfDNA testing provided results in 249 (96.1%). Further, cfDNA testing identified 97.2% (35/36) of trisomy 21, 100% (13/13) of trisomy 18, 100% of trisomy 13 (5/5), and 75% of sex chromosome aneuploidies (3/4). The authors conclude that “a policy of performing an invasive test in [individuals] with a combined risk of ≥1 in 10 or NT ≥4 mm and offering cfDNA testing to the remaining cases would detect all cases of trisomy 21, 18 or 13, 80% of sex aneuploidies and 62.5% of other defects and would avoid an invasive procedure in 82.4% of euploid fetuses.”19 These data support the earlier meta-analysis that reported NIPT sensitivity of trisomy 21, trisomy 18, and trisomy 13 of 99%, 96.8%, and 92.1%, respectively and specificities of 99.92%, 99.85%, and 99.80%, respectively, for trisomies 21, 18, and 13.20,21
A multi-year study of more than 5000 patients in public hospitals in Spain examined the effect of NIPT on the number of invasive procedures performed, showing that the introduction of NIPT drastically reduced the incidences of invasive procedures. The data shows that despite a 60.5% reduction occurred in invasive procedures, the chromosomopathy detection rate was unaffected; moreover, the ratio of positive invasive procedures was improved to 50%, indicating that unwarranted invasive procedures had been avoided.22 The authors of the study concluded, “NIPT introduction has caused a significant reduction of 60.5% of IP [invasive procedures] in high chromosomopathy risk patients after combined screening without modifying detection rate.”22
A meta-analysis was completed by Mackie, et al. (2017), researching the accuracy of cell-free fetal DNA NIPT testing in singleton pregnancies. A total of 117 studies were included, analyzing 18 different conditions. For RHD testing, a sensitivity of 0.993 and specificity of 0.984 was identified and for fetal sex identification, a sensitivity of 0.989 and a specificity of 0.996 was calculated.23 With such high sensitivity and specificity calculations, NIPT testing for fetal sex and RHD status may be considered accurate diagnostic tools.
Clausen, et al. (2014) completed a two-year evaluation of nationwide prenatal RhD screening in Denmark. A total of 12,668 pregnancies were analyzed, with blood samples drawn in week 25 of pregnancy. DNA was extracted from these blood samples and was analyzed for the RHD gene. Results were later compared to the serological typing of the newborns after birth. “The sensitivity for the detection of fetal RHD was 99.9% (95% CI: 99.7-99.9%). Unnecessary recommendation of prenatal RhD prophylaxis was avoided in 97.3% of the [individuals] carrying an RhD-negative fetus. Fetuses that were seropositive for RhD were not detected in 11 pregnancies (0.087%).”24 This study shows high sensitivity of fetal RHD genotyping, results which were recently supported by another large-scale meta-analysis completed by Yang, et al. (2019), focusing on NIPT testing for fetal RhD status. A total of 3921 results confirmed that “High-throughput NIPT is sufficiently accurate to detect fetal RhD status in RhD-negative [individuals] and would considerably reduce unnecessary treatment with routine anti-D immunoglobulin.”25
Darlington, et al. (2018) completed an analysis of 11 French Obstetric Departments with a total of 949 patients to determine the effectiveness of RhD genotyping. The patients were separated into two groups (genotyping group: n=515, and control group: n=335). The authors concluded that “Early knowledge of the RHD status of the fetus using non-invasive fetal RHD genotyping significantly improved the management of RHD negative pregnancies with a small increase in cost.”26
Runkel, et al. (2020) completed a systematic review to determine the benefit of NIPT for fetal RhD status in RhD-negative pregnant individuals because “All non-sensitized Rhesus D (RhD)-negative pregnant [individuals] in Germany receive antenatal anti-D prophylaxis without knowledge of fetal RhD status.” The meta-analysis included data from 60,000 participants, with the focus of the research on the impact of fetal and maternal morbidity. The researchers concluded that “NIPT for fetal RhD status is equivalent to conventional serologic testing using the newborn’s blood. Studies investigating patient-relevant outcomes are still lacking.”27
Hoskovec, et al. (2023) evaluated the “clinical performance of carrier screening for cystic fibrosis, hemoglobinopathies, and spinal muscular atrophy with reflex single-gene noninvasive prenatal screening (sgNIPS).”7 In the study, 9151 pregnant individuals were screened for carrier status. As a result, 1669 (18.2%) of the sampled individuals were found to carry one or more harmful genetic variations and were subsequently tested using sgNIPS. The results of sgNIPS were then compared to the outcomes of 201 pregnancies, which were obtained from surveys completed by parents or reports from healthcare providers. In conclusion, carrier screening using sgNIPS during pregnancy presents an alternative approach that circumvents the need for a paternal sample. It offers accurate assessment of fetal risk promptly, facilitating prenatal counseling and pregnancy management.7
Westin, et al. (2022) conducted a retrospective study which aimed to “validate the sgNIPT in clinical samples and identify high-risk SCD fetuses in a cohort of at-risk pregnancies.” This retrospective clinical investigation gathered 77 maternal blood samples from pregnant patients at either Baylor College of Medicine or the University of Alabama at Birmingham. These patients were identified as having at least one harmful HBB allele. The results of this study highlighted that sgNIPT screening promotes “efficient and accurate fetal risk assessment for SCD in pregnant patients.”28
It is notable that the field continues to evolve, with potential shifts from one testing method to another in pursuit of optimality and comprehensiveness. A multicenter retrospective study of singleton high-risk pregnancies for chromosomal abnormalities was conducted by Zhu, et al. (2020) to evaluate the utility of expanded non-invasive prenatal screening as compared with chromosomal microarray analysis (CMA). The analysis enrolled subjects who underwent expanded NIPS and CMA sequentially during pregnancy from 2015 through 2019. The study demonstrated that of the 943 high‐risk pregnancies, 550 (58.3%) cases had positive NIPS results, while positive CMA results were detected in 308 (32.7%) cases, and the agreement rates between NIPS and CMA were 82.3%, 59.6% and 25.0% for trisomy 21, 18 and 13, respectively. Regarding rare aneuploidies and segmental imbalances, NIPS and CMA results were concordant in 7.5% and 33.3% of cases. However, copy number variants were better detected with CMA than with NIPS and additional genetic aberrations were detected by CMA in one of 17 high-risk pregnancies that were otherwise passed over when processed with NIPS. The researchers contend that CMA should be offered for high‐risk pregnancies to provide comprehensive detection of chromosomal abnormalities in these pregnancies.29
Zhang, et al. (2025) studied the clinical utility of plasma cfDNA and NGS based NIPT for the detection of dominant single-gene disorders (dSGDs). The authors used NIPT methods to target 34 genes, with 25 genes specifically correlated to neurodevelopmental disorders. Testing was performed on 567 pregnant people. The results were compared to invasive prenatal or postnatal genetic diagnosis obtained using whole-exome sequencing and Sanger sequencing. “NIPT-dSGD did not generate any false-positive or negative results, achieving 100% of sensitivity (95% CI, 71.7%-100%) and 100% of specificity (95% CI, 99.0%-100%).” The authors concluded that “NIPT-dSGD provides accurate genetic testing for de novo and paternally inherited variants of dominant genes, including those that do not cause any ultrasound abnormalities, which could assist clinicians and families in better pregnancy management.”30
This policy focuses on genetic testing performed during pre-conception and/or prenatal periods as part of a comprehensive prenatal care program.
American College of Medical Genetics and Genomics (ACMG)
In 2021, ACMG released an updated guideline for screening for autosomal recessive and X-linked conditions during pregnancy and pre-conception. Their practice resource reviews aim to recommend “a consistent and equitable approach for offering carrier screening to all individuals during pregnancy and preconception” and replaces any earlier ACMG position statements on prenatal/pre-conception expanded carrier screening and provide the following recommendations:
- “Analytical validity of carrier screening is to be established by a laboratory in compliance with CLIA/CAP regulations and adhering to ACMG Laboratory Standards and Guidelines.”
- “As evidence evolves, ClinVar and ClinGen continually update pathogenicity of variants and the association between genes and conditions, respectively.”
- “Carrier screening enables those screened to consider their reproductive risks, reproductive options, and to make informed decisions.”
- “Published evidence supports clinical utility for carrier screening of multiple conditions simultaneously.”
- “The phrase “expanded carrier screening” be replaced by “carrier screening.”
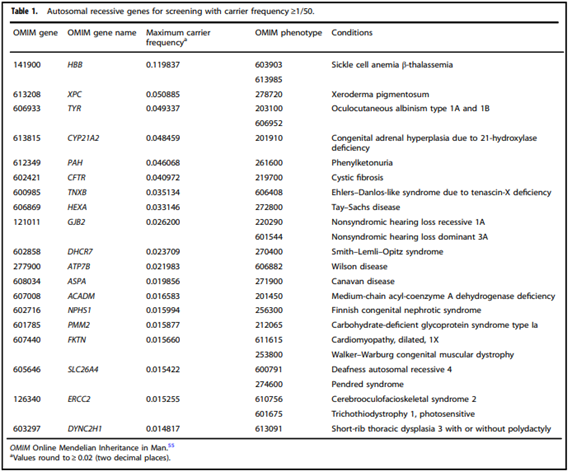
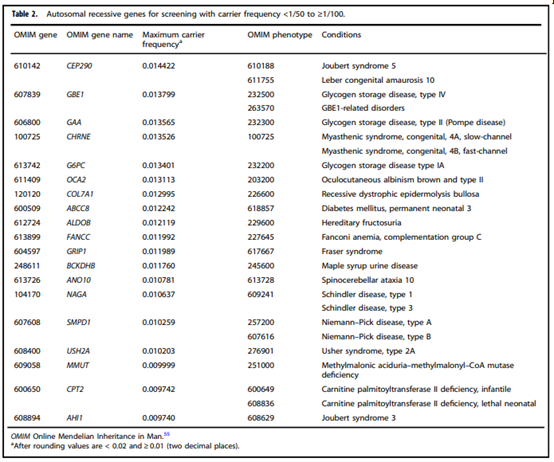
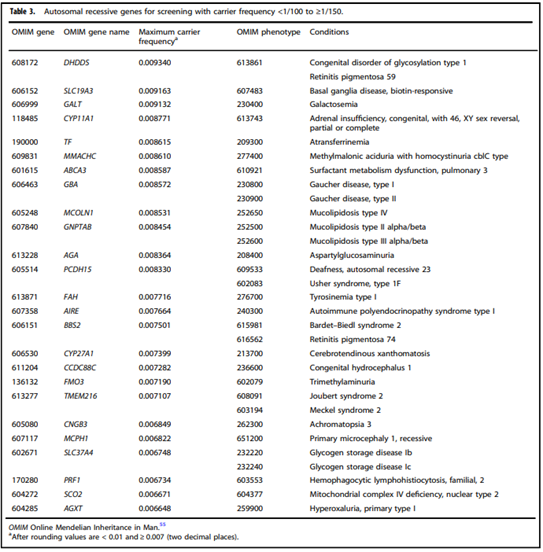
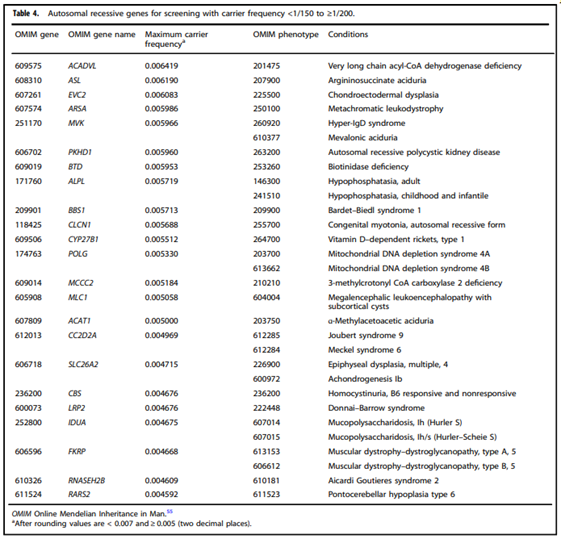
- “Adopting a more precise tiered system based on carrier frequency:
- Tier 4: <1/200 carrier frequency (includes Tier 3) genes/condition will vary by lab
- Tier 3: ≥ 1/200 carrier frequency (includes Tier 2) includes X-linked conditions
- Tier 2: ≥1/100 carrier frequency (includes Tier 1)
- Tier 1: CF [Cystic Fibrosis] + SMA [spinal muscular atrophy] + Risk Based Screening”
- “Tier 1 screening conveys the recommendations previously adopted by ACMG and ACOG” and “adopts an ethnic and population neutral approach when screening for cystic fibrosis and spinal muscular atrophy. Beyond these two conditions, additional carrier screening is determined after risk assessment, which incorporates personal medical and family history as well as laboratory and imaging information where appropriate.”
- “Tier 2 carrier screening stems from an ACOG recommendation for conditions that have a severe or moderate phenotype and a carrier frequency of at least 1/100.” However, “data demonstrate that carrier screening for two common conditions using a carrier frequency threshold of 1/100 may not be equitable across diverse populations. Others have shown that limiting the carrier frequency to ≥1/100 creates missed opportunities to identify couples at risk for serious conditions.”
- “We define Tier 3 screening as carrier screening for conditions with a carrier frequency ≥1/200 . . . Tier 2 and Tier 3 screening prioritize carrier frequency as a way to think about conditions most appropriate for screening in the general population. However, when ACOG proposed this level, they did not specify whether it was thinking about carrier frequency in terms of the global population or subpopulations. We use “carrier frequency” to mean in any ethnic group with reasonable representation in the United States.”
- “Tier 4 includes genes less common than those in Tier 3 and can identify additional at-risk couples. Tier 4 has no lower limit carrier screening frequency and can greatly extend the number of conditions screened . . . the clinical validity at this level of carrier screening may be less compelling, therefore we suggest reserving this level of screening for consanguineous pregnancies (second cousins or closer) and in couples where family or medical history suggests Tier 4 screening might be beneficial . . . Importantly, patients should understand that their chance of being a carrier for one or more conditions increases as the number of conditions screened is increased.”
- “All pregnant patients and those planning a pregnancy should be offered Tier 3 carrier screening.
- Tier 4 screening should be considered:
- When a pregnancy stems from a known or possible consanguineous relationship (second cousins or closer).
- When a family or personal medical history warrants”
- ACMG does NOT recommend:
- Offering Tier 1 and/or Tier 2 screening, because these do not provide equitable evaluation of all racial/ethnic groups.
- Routine offering of Tier 4 panels.”
- “Carrier screening paradigms should be ethnic and population neutral and more inclusive of diverse populations to promote equity and inclusion.”
- “All pregnant patients and those planning a pregnancy should be offered Tier 3 carrier screening for autosomal recessive (Tables 1–5) and X-linked (Table 6) conditions.”
- “Reproductive partners of pregnant patients and those planning a pregnancy may be offered Tier 3 carrier screening for autosomal recessive conditions (Tables 1–5) when carrier screening is performed simultaneously with their partner.”
- “All XX patients should be offered screening for only those X-linked genes listed in Table 6 as part of Tier 3 screening.”
- “When Tier 1 or Tier 2 carrier screening was performed in a prior pregnancy, Tier 3 screening should be offered.”31
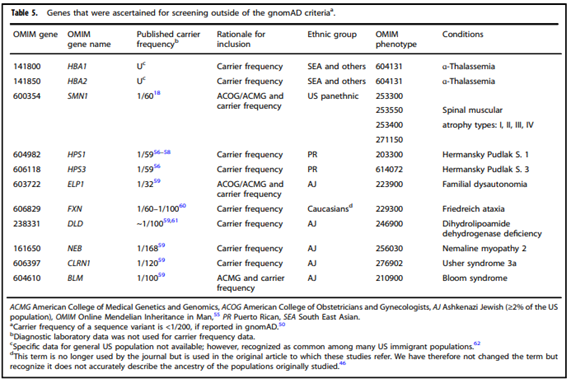
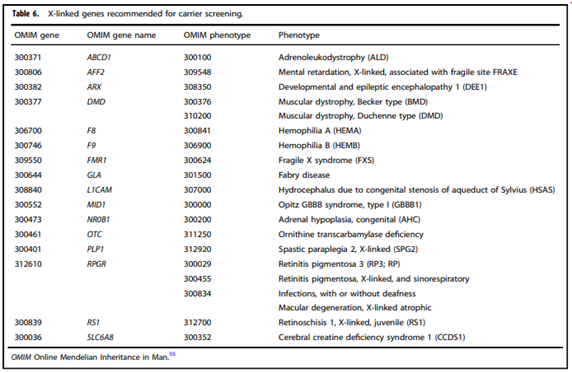
CFTR Variant Testing
In 2020, the ACMG provided a technical standard for CFTR variant testing. These standards state the following as it pertains to pregnancy:
“During pregnancy, simultaneous testing may be desired depending on gestational age, family and personal history, ethnicity, or patient preferences. Carrier testing may be offered to individuals with a positive family history of CF, in partners of individuals with a positive family history, in partners of CAVD males, to reproductive age women, and to gamete donors. CFTR variant testing can also be performed for prenatal diagnosis using cells obtained for diagnostic cytogenetic testing (i.e., amniocentesis or chorionic villus sampling [CVS]).”32
“As a way to ensure that CFTR variant testing for carrier screening and diagnostic testing purposes remains inclusive, the ACMG recommends either a classification-based reporting approach or a classification-based (targeted) testing approach (which has historically been used for CFTR carrier screening). For those laboratories who wish to continue using a targeted testing approach, the ACMG-23 variant panel remains as the minimum list of CFTR variants that should be included. Laboratories may want to consider adding additional variants to their panel depending on the ethnic composition of their expected test population. However, the minimum list of CFTR variants recommended for pan-ethnic carrier screening has not been increased at this time.”32
In 2023, the ACMG provided updated recommendations for CFTR carrier screening which includes a new minimum CFTR variant set (increased from 23 to 100 variants). The updated ACMG position statement states the following:
“This new set now supersedes the previous set of 23 CFTR variants recommended by the ACMG. These revised recommendations apply only to carrier screening. They do not apply to CFTR variant testing for diagnosis or newborn screening. All other aspects of the updated 2020 ACMG CFTR technical standards still apply.”32,33
American College of Obstetricians and Gynecologists (ACOG)
ACOG has several practice guidelines related to prenatal care as well as both pre-conception and prenatal testing. ACOG recommendations and guidelines include the following:
Genetic Testing and Genetic Counseling: Concerning genetic testing and genetic counseling, ACOG recommends:
- “The routine use of whole-genome or whole-exome sequencing for prenatal diagnosis is not recommended outside of the context of clinical trials until sufficient peer-reviewed data and validation studies are published.”4 This was reaffirmed in 2023.
- Chromosomal microarray analysis (CMA) is recommended for patients with a fetus with at least one major structure abnormality identified via ultrasound. CMA can be considered for all pregnant individuals who undergo prenatal diagnostic testing; however, “In a patient with a structurally normal fetus who is undergoing invasive prenatal diagnostic testing, either fetal karyotyping or a chromosomal microarray analysis can be performed. Chromosomal microarray analysis of fetal tissue (ie, amniotic fluid, placenta, or products of conception) is recommended in the evaluation of intrauterine fetal death or stillbirth when further cytogenetic analysis is desired because of the test’s increased likelihood of obtaining results and improved detection of causative abnormalities.”4 This was reaffirmed in 2023.
- “All patients who are considering pregnancy or are already pregnant, regardless of screening strategy and ethnicity, should be offered carrier screening for cystic fibrosis and spinal muscular atrophy, as well as a complete blood count and screening for thalassemias and hemoglobinopathies. Fragile X premutation carrier screening is recommended for [individuals] with a family history of fragile X-related disorders or intellectual disability suggestive of fragile X syndrome, or [individuals] with a personal history of ovarian insufficiency. Additional screening also may be indicated based on family history or specific ethnicity.”34 This was reaffirmed in 2023.
- “The American College of Obstetricians and Gynecologists discourages direct-to-consumer genetic testing without appropriate counseling. . . Patients may present after direct-to-consumer testing already has been performed, and clinicians should be prepared to review these results or refer to a health care professional with the appropriate knowledge, training, and experience in interpreting test results. . . Given the insufficient data to support the use of single nucleotide polymorphisms (SNP) testing for medical purposes, SNP testing to provide individual risk assessment for a variety of diseases or to tailor drug therapy outside of an institutional review board-approved research protocol is not recommended. The American College of Obstetricians and Gynecologists recommends that the use of these technologies be viewed as investigational at this time.”35 This was reaffirmed in 2023.
- ACOG notes that “Cascade testing has been shown to be cost effective in part because testing for specific mutations (e.g., those identified in the affected relative) is less expensive than whole-gene sequencing.”36 This was reaffirmed in 2022.
Prenatal Diagnostic Testing for Genetic Disorders: Concerning prenatal diagnostic testing for genetic disorders, ACOG has published the following recommendations:
- “An abnormal FISH result should not be considered diagnostic. Therefore, clinical decision making based on information from FISH should include at least one of the following additional results: confirmatory traditional metaphase chromosome analysis or chromosomal microarray, or consistent clinical information (such as abnormal ultrasonographic findings or a positive screening test result for Down syndrome or trisomy 18).”
- “All pregnant women should be offered prenatal assessment for aneuploidy by screening or diagnostic testing regardless of maternal age or other risk factors.”
- “Prenatal genetic testing cannot identify all abnormalities or problems in a fetus, and any testing should be focused on the individual patient’s risks, reproductive goals and preferences.”
- “Genetic testing should be discussed as early as possible in pregnancy, ideally at the first obstetric visit, so that first-trimester options are available.”37
Prevention of Rh D Alloimmunization: Concerning the prevention of Rh D alloimmunization, ACOG has published the guidelines supporting the administration of anti-D immune globulin to individuals in various scenarios. However, these guidelines do not mention the use of cell-free fetal DNA for fetal RHD testing to determine if anti-D immune globulin is needed.38
Paternal and Fetal Genotyping in the Management of Alloimmunization in Pregnancy:39
- “Paternal RHD zygosity testing using genotypic analysis is recommended for Rh-D alloimmunization risk assessment. It may be reasonable to defer or discontinue fetal surveillance for anemia in the setting of paternal genotyping that is RHD homozygous negative.”
- “Fetal antigen genotyping is recommended when the paternal genotype is heterozygous or unknown.”
- “Because cfDNA testing possesses performance characteristics that appear comparable with those of molecular testing, while avoiding the rare complications and costs associated with diagnostic genetic testing, it is reasonable to use it as an alternative tool for fetal RHD testing among alloimmunized patients with potentially at-risk pregnancies who decline amniocentesis.”
- “Cell-free DNA for the assessment of selected non–Rh-D red blood cell antigens may be considered for pregnant patients declining amniocentesis, after weighing cost, access, and the encouraging-yet-limited data supporting its use.”
In case of human Rho(D) immune globulin (Rhlg) shortages, ACOG has “prioritization and conservation strategies for consideration.” “Although current ACOG guidance does not recommend routine use of noninvasive prenatal testing (NIPT) to determine fetal Rh(D) status based on cost-effectiveness analyses, the use of NIPT to prioritize use of RhIg and conserve RhIg supply is a reasonable consideration in the practice setting that is experiencing RhIg shortages.” ACOG further states that “Noninvasive fetal red blood cell antigen genotyping utilizing cell-free DNA (cfDNA) isolated from maternal plasma has demonstrated high sensitivity and specificity for detection of fetal Rh(D) antigen status. If cfDNA testing results confirm an Rh(D)-negative fetus, RhIg would not need to be routinely administered in the antepartum period (for bleeding, abortion, pregnancy loss, or at 28 weeks of gestation). Available cfDNA testing options for Rh(D) may vary depending on location and practice setting (eg, companies offering the test; whether the test is offered as a stand-alone or combined with aneuploidy testing; timing of results; insurance coverage) and should be confirmed before implementation.”40
Genetic Carrier Screening: Concerning genetic carrier screening, including testing for specific conditions, ACOG recommends (reaffirmed 2023):34,41
- “Carrier screening and counseling ideally should be performed before pregnancy.
- “If an individual is found to be a carrier for a specific condition, the individual’s reproductive partner should be offered testing in order to receive informed genetic counseling about potential reproductive outcomes. Concurrent screening of the patient and her partner is suggested if there are time constraints for decisions about prenatal diagnostic evaluation.”
- “Carrier screening for a particular condition generally should be performed only once in a person’s lifetime, and the results should be documented in the patient’s health record. Because of the rapid evolution of genetic testing, additional mutations may be included in newer screening panels. The decision to rescreen a patient should be undertaken only with the guidance of a genetics professional who can best assess the incremental benefit of repeat testing for additional mutations.”
- “Prenatal carrier screening does not replace newborn screening, nor does newborn screening replace the potential value of prenatal carrier screening.”
- “The cost of carrier screening for an individual condition may be higher than the cost of testing through commercially available expanded carrier screening panels. When selecting a carrier screening approach, the cost of each option to the patient and the health care system should be considered.”
- “Screening for spinal muscular atrophy should be offered to all [individuals] who are considering pregnancy or are currently pregnant. In patients with a family history of spinal muscular atrophy, molecular testing reports of the affected individual and carrier testing of the related parent should be reviewed, if possible, before testing. If the reports are not available, SMN1 deletion testing should be recommended for the low-risk partner.”
- “Cystic fibrosis carrier screening should be offered to all [individuals] who are considering pregnancy or are currently pregnant. Complete analysis of the CFTR gene by DNA sequencing is not appropriate for routine carrier screening.”
- “A complete blood count with red blood cell indices should be performed in all [individuals] who are currently pregnant to assess not only their risk of anemia but also to allow assessment for risk of a hemoglobinopathy. Ideally, this testing also should be offered to [individuals] before pregnancy. A hemoglobin electrophoresis should be performed in addition to a complete blood count if there is suspicion of hemoglobinopathy based on ethnicity (African, Mediterranean, Middle Eastern, Southeast Asian, or West Indian descent). If red blood cell indices indicate a low mean corpuscular hemoglobin or mean corpuscular volume, hemoglobin electrophoresis also should be performed.”
- “Fragile X premutation carrier screening is recommended for [individuals] with a family history of fragile X-related disorders or intellectual disability suggestive of fragile X syndrome and who are considering pregnancy or are currently pregnant.”
- “If a [individual] has unexplained ovarian insufficiency or failure or an elevated follicle-stimulating hormone level before age 40 years, fragile X carrier screening is recommended to determine whether she has an FMR1 premutation.”
- “All identified individuals with intermediate results and carriers of a fragile X premutation or full mutation should be provided follow-up genetic counseling to discuss the risk to their offspring of inheriting an expanded full-mutation fragile X allele and to discuss fragile X-associated disorders (premature ovarian insufficiency and fragile X tremor/ataxia syndrome).”
- “Prenatal diagnostic testing for fragile X syndrome should be offered to known carriers of the fragile X premutation or full mutation.”
- “DNA-based molecular analysis (eg, Southern blot analysis and polymerase chain reaction) is the preferred method of diagnosis of fragile X syndrome and of determining FMR1 triplet repeat number (e.g., premutations). In rare cases, the size of the triplet repeat and the methylation status do not correlate, which makes it difficult to predict the clinical phenotype. In cases of this discordance, the patient should be referred to a genetics professional.”
- “When only one partner is of Ashkenazi Jewish descent, that individual should be offered screening first. If it is determined that this individual is a carrier, the other partner should be offered screening. However, the couple should be informed that the carrier frequency and the detection rate in non-Jewish individuals are unknown for most of these disorders, except for Tay–Sachs disease and cystic fibrosis. Therefore, it is difficult to accurately predict the couple’s risk of having a child with the disorder.”
- “Screening for Tay–Sachs disease should be offered when considering pregnancy or during pregnancy if either member of a couple is of Ashkenazi Jewish, French–Canadian, or Cajun descent. Those with a family history consistent with Tay–Sachs disease also should be offered screening. When one member of a couple is at high risk (i.e., of Ashkenazi Jewish, French–Canadian, or Cajun descent or has a family history consistent with Tay–Sachs disease) but the other partner is not, the high-risk partner should be offered screening. If the high-risk partner is found to be a carrier, the other partner also should be offered screening.”
- “Enzyme testing in pregnant [individuals] and [individuals] taking oral contraceptives should be performed using leukocyte testing because serum testing is associated with an increased false-positive rate in these populations.”
- “If Tay–Sachs disease screening is performed as part of pan-ethnic expanded carrier screening, it is important to recognize the limitations of the mutations screened in detecting carriers in the general population. In the presence of a family history of Tay–Sachs disease, expanded carrier screening panels are not the best approach to screening unless the familial mutation is included on the panel.”41
- Regarding expanded carrier screening panels, ACOG recommends that “the disorders selected for inclusion should meet several of the following consensus-determined criteria: have a carrier frequency of 1 in 100 or greater, have a well-defined phenotype, have a detrimental effect on quality of life, cause cognitive or physical impairment, require surgical or medical intervention, or have an onset early in life.” ACOG further states that “screened conditions should be able to be diagnosed prenatally and may afford opportunities for antenatal intervention to improve perinatal outcomes, changes to delivery management to optimize newborn and infant outcomes, and education of the parents about special care needs after birth.”34
Carrier Screening in the Age of Genomic Medicine: Concerning carrier screening in the age of genomic medicine, the ACOG has published the following guidelines:34
- “Ethnic-specific, pan-ethnic and expanded carrier screening are acceptable strategies for prepregnancy and prenatal carrier screening.
- If a patient requests a screening strategy other than the one used by the obstetrician-gynecologist or other health care provider, the requested test should be made available to her after counseling on its limitations, benefits, and alternatives.
- All patients who are considering pregnancy or already pregnant, regardless of screening strategy and ethnicity, should be offered carrier screening for cystic fibrosis and spinal muscular atrophy, as well as a complete blood count and screening for thalassemias and hemoglobinopathies. Fragile X premutation carrier screening is also recommended for [individuals] with a family history of fragile x-related disorders or intellectual disability suggestive of fragile X syndrome, or [individuals] with a personal history of ovarian insufficiency. Additional screening also may be indicated based on family history or specific ethnicity.
- If a [individual] is found to be a carrier for a specific condition, her reproductive partner should be offered screening to provide accurate genetic counseling for the couple with regard to the risk of having an affected child. Additional genetic counseling should be provided to discuss the specific condition, residual risk, and options for prenatal testing.
- Individuals with a family history of a genetic disorder may benefit from the identification of the specific familial mutation or mutations rather than carrier screening. Knowledge of the specific familial mutation may allow for more specific and rapid prenatal diagnosis.
- Given the multitude of conditions that can be included in expanded carrier screening panels, the disorders selected for inclusion should meet several of the following consensus-determined criteria: have a carrier frequency of 1 in 100 or greater, have a well-defined phenotype, have a detrimental effect on quality of life, cause cognitive or physical impairment, require surgical or medical intervention, or have an onset early in life. Additionally, screened conditions should be able to be diagnosed prenatally and may afford opportunities for antenatal intervention to improve perinatal outcomes, changes to delivery management to optimize newborn and infant outcomes, and education of the parents about special care needs after birth.
- Carrier screening panels should not include conditions primarily associated with a disease of adult onset.”34 This guideline was reaffirmed in 2023.
International Society for Prenatal Diagnosis (ISPD), the Society for Maternal Fetal Medicine (SMFM), and the Perinatal Quality Foundation (PQF)
The ISPD, SMFM and PQF published the following guidelines on the use of genome-wide sequencing for fetal diagnosis:
- The use of diagnostic sequencing is currently being introduced for evaluation of fetuses for whom standard diagnostic genetic testing, such as chromosomal microarray analysis (CMA), has already been performed and is uninformative, is offered concurrently according to accepted practice guidelines, or for whom expert genetic opinion determines that standard genetic testing is less optimal than sequencing for the presenting fetal phenotype.
- The routine use of prenatal sequencing as a diagnostic test cannot currently be supported due to insufficient validation data and knowledge about its benefits and pitfalls.42
In addition to the joint position statement released in 2018, the IPSD released a guideline in 2020 on the use of cfDNA screening for trisomies in multiple pregnancies:
- “The use of first trimester cfDNA screening for the common autosomal trisomies is appropriate for twin pregnancies due to sufficient evidence showing high detection and low false positive rates with high predictive values. Moderate.”
- “It is preferable for laboratories performing cfDNA testing in multi-fetal pregnancies to take evidence of zygosity into consideration (eg, chorionicity, sex of the fetuses, embryo transfer history) for the interpretation of both test results and fetal fractions. Moderate.”
- “Screening options for triplet pregnancies are lacking and cfDNA may be a potential option. However, diagnostic testing should always be offered and the limitations of screening tests stressed. Low.”43
College of American Pathologists (CAP) Transfusion Medicine Resource Committee (TMRC) Work Group
The following recommendations were given by the CAP TMRC Work Group:
- The Work Group recommends that RHD genotyping be performed whenever a discordant RhD typing result and/or a serological weak D phenotype is detected in patients, including pregnant individuals, newborns, and potential transfusion recipients. It is anticipated that the immediate benefit will be fewer unnecessary injections of RhIG and increased availability of RhD-negative RBCs for transfusion.
- Other than RHD genotypes weak D type 1, 2, or 3, the Work Group recommends that individuals with a serological weak D phenotype receive conventional prophylaxis with RhIG, including postpartum RhIG if the newborn is RhD-positive or has a serological weak D phenotype.44
British Society for Haematology (BSH)
The BSH states that, “Invasive testing is not contraindicated if alloimmunisation has occurred. There is a risk of increasing the degree of alloimmunisation, or causing alloimmunisation to new antigens, as a result of the procedure, and this should be discussed with the woman. If the mother is RhD negative and is not already sensitised, anti-D immunoglobulin should be given following the procedure.” The BSH also reiterates that “The use of high- throughput, non-invasive prenatal diagnosis of foetal RHD status was recommended by the National Institute for Health and Care Excellence (NICE) in 2016.”45
References
1. Grant A, Mohide P. Screening and diagnostic tests in antenatal care. Effectiveness and satisfaction in antenatal care. 1982:22-59.
2. Lockwood CJ, Magriples U. Prenatal care: Initial assessment. Wolters Kluwer. Updated May 15. https://www.uptodate.com/contents/prenatal-care-initial-assessment
3. de Jong A, Maya I, van Lith JM. Prenatal screening: current practice, new developments, ethical challenges. Bioethics. 2015;29(1):1-8. doi:10.1111/bioe.12123
4. ACOG. Committee Opinion No.682: Microarrays and Next-Generation Sequencing Technology: The Use of Advanced Genetic Diagnostic Tools in Obstetrics and Gynecology. Obstetrics and gynecology. Dec 2016;128(6):e262-e268. doi:10.1097/aog.0000000000001817
5. Miller D. Use of chromosomal microarray in obstetrics. Updated July 31, 2024. https://www.uptodate.com/contents/use-of-chromosomal-microarray-in-obstetrics
6. LabCorp. Inheritest. https://www.integratedgenetics.com/patients/pre-pregnancy/inheritest
7. Hoskovec J, Hardisty EE, Talati AN, et al. Maternal carrier screening with single-gene NIPS provides accurate fetal risk assessments for recessive conditions. Genetics in Medicine. 2023;25(2):100334. doi:10.1016/j.gim.2022.10.014
8. BillionToOne. UNITY Complete: The New Standard In Prenatal Care. https://www.unityscreen.com/
9. Calhoun D, Marvin Bahr T. Alloimmune hemolytic disease of the newborn: Postnatal diagnosis and management. Updated July 15, 2024. https://www.uptodate.com/contents/alloimmune-hemolytic-disease-of-the-newborn-postnatal-diagnosis-and-management
10. Moise K. RhD alloimmunization: Prevention in pregnant and postpartum patients. Updated September 17, 2024. https://www.uptodate.com/contents/rhd-alloimmunization-prevention-in-pregnant-and-postpartum-patients
11. Daniels G, Finning K, Martin P, Summers J. Fetal RhD genotyping: a more efficient use of anti-D immunoglobulin. Transfus Clin Biol. 2007;14(6):568-71. doi:10.1016/j.tracli.2008.03.007
12. Kent J, Farrell AM, Soothill P. Routine administration of Anti-D: the ethical case for offering pregnant women fetal RHD genotyping and a review of policy and practice. BMC Pregnancy Childbirth. 2014;14:87. doi:10.1186/1471-2393-14-87
13. Finning K, Martin P, Summers J, Massey E, Poole G, Daniels G. Effect of high throughput RHD typing of fetal DNA in maternal plasma on use of anti-RhD immunoglobulin in RhD negative pregnant women: prospective feasibility study. Bmj. 2008;336(7648):816-8. doi:10.1136/bmj.39518.463206.25
14. de Haas M, Thurik FF, van der Ploeg CP, et al. Sensitivity of fetal RHD screening for safe guidance of targeted anti-D immunoglobulin prophylaxis: prospective cohort study of a nationwide programme in the Netherlands. Bmj. Nov 7 2016;355:i5789. doi:10.1136/bmj.i5789
15. Manfroi S, Calisesi C, Fagiani P, et al. Prenatal non-invasive foetal RHD genotyping: diagnostic accuracy of a test as a guide for appropriate administration of antenatal anti-D immunoprophylaxis. Blood Transfus. 2018;16(6):514-524. doi:10.2450/2018.0270-17
16. Yesilcinar I, Guvenc G. Counselling and education for prenatal screening and diagnostic tests for pregnant women: Randomized controlled trial. Int J Nurs Pract. 2021;27(5):e13000. doi:10.1111/ijn.13000
17. Migliorini S, Saccone G, Silvestro F, et al. First-trimester screening based on cell-free DNA vs combined screening: A randomized clinical trial on women's experience. Prenat Diagn. 2020;40(11):1482-1488. doi:10.1002/pd.5800
18. Biro O, Rigo J, Jr., Nagy B. Noninvasive prenatal testing for congenital heart disease - cell-free nucleic acid and protein biomarkers in maternal blood. J Matern Fetal Neonatal Med. Mar 2020;33(6):1044-1050. doi:10.1080/14767058.2018.1508437
19. Persico N, Boito S, Ischia B, et al. Cell‐free DNA testing in the maternal blood in high‐risk pregnancies after first‐trimester combined screening. Prenatal Diagnosis. 2016;36(3):232-236.
20. Dondorp W, de Wert G, Bombard Y, et al. Non-invasive prenatal testing for aneuploidy and beyond: challenges of responsible innovation in prenatal screening. European journal of human genetics : EJHG. 2015;23(11):1438-1450. doi:10.1038/ejhg.2015.57
21. Gil MM, Akolekar R, Quezada MS, Bregant B, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: meta-analysis. Fetal diagnosis and therapy. 2014;35(3):156-73. doi:10.1159/000358326
22. Martinez-Payo C, Bada-Bosch I, Martinez-Moya M, Perez-Medina T. Clinical results after the implementation of cell-free fetal DNA detection in maternal plasma. The journal of obstetrics and gynaecology research. 2018;44(8):1369-1376. doi:10.1111/jog.13672
23. Mackie FL, Hemming K, Allen S, Morris RK, Kilby MD. The accuracy of cell-free fetal DNA-based non-invasive prenatal testing in singleton pregnancies: a systematic review and bivariate meta-analysis. Bjog. 2017;124(1):32-46. doi:10.1111/1471-0528.14050
24. Clausen FB, Steffensen R, Christiansen M, et al. Routine noninvasive prenatal screening for fetal RHD in plasma of RhD-negative pregnant women-2 years of screening experience from Denmark. Prenat Diagn. 2014;34(10):1000-5. doi:10.1002/pd.4419
25. Yang H, Llewellyn A, Walker R, et al. High-throughput, non-invasive prenatal testing for fetal rhesus D status in RhD-negative women: a systematic review and meta-analysis. BMC Med. Feb 14 2019;17(1):37. doi:10.1186/s12916-019-1254-4
26. Darlington M, Carbonne B, Mailloux A, et al. Effectiveness and costs of non-invasive foetal RHD genotyping in rhesus-D negative mothers: a French multicentric two-arm study of 850 women. BMC Pregnancy Childbirth. 2018;18(1):496. doi:10.1186/s12884-018-2114-5
27. Runkel B, Bein G, Sieben W, Sow D, Polus S, Fleer D. Targeted antenatal anti-D prophylaxis for RhD-negative pregnant women: a systematic review. BMC Pregnancy Childbirth. 2020;20(1):83. doi:10.1186/s12884-020-2742-4
28. Westin ER, Tsao DS, Atay O, et al. Validation of single‐gene noninvasive prenatal testing for sickle cell disease. American Journal of Hematology. 2022;97(7)doi:10.1002/ajh.26570
29. Zhu X, Chen M, Wang H, et al. Clinical utility of expanded noninvasive prenatal screening and chromosomal microarray analysis in high risk pregnancies. Ultrasound Obstet Gynecol. 2020;doi:10.1002/uog.22021
30. Zhang H, He J, Teng Y, et al. Non-invasive prenatal testing for dominant single-gene disorders using targeted next-generation sequencing. Qjm. Feb 21 2025;doi:10.1093/qjmed/hcaf017
31. Gregg AR, Aarabi M, Klugman S, et al. Screening for autosomal recessive and X-linked conditions during pregnancy and preconception: a practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2021;23(10):1793-1806. doi:10.1038/s41436-021-01203-z
32. Deignan JL, Astbury C, Cutting GR, et al. CFTR variant testing: a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine. 2020/08/01/ 2020;22(8):1288-1295. doi:10.1038/s41436-020-0822-5
33. Deignan JL, Gregg AR, Grody WW, et al. Updated recommendations for CFTR carrier screening: A position statement of the American College of Medical Genetics and Genomics (ACMG). Genetics in Medicine. 2023;25(8):100867. doi:10.1016/j.gim.2023.100867
34. ACOG. Committee Opinion No. 690: Carrier Screening in the Age of Genomic Medicine. Obstetrics and gynecology. Mar 2017;129(3):e35-e40. doi:10.1097/aog.0000000000001951
35. ACOG. Consumer Testing for Disease Risk: ACOG Committee Opinion, Number 816. Obstetrics and gynecology. Jan 1 2021;137(1):e1-e6. doi:10.1097/AOG.0000000000004200
36. ACOG. ACOG Committee Opinion No. 727: Cascade Testing: Testing Women for Known Hereditary Genetic Mutations Associated With Cancer. Obstetrics and gynecology. Jan 2018;131(1):e31-e34. doi:10.1097/aog.0000000000002457
37. ACOG. Practice Bulletin No. 162: Prenatal Diagnostic Testing for Genetic Disorders. Obstetrics and gynecology. May 2016;127(5):e108-e122. doi:10.1097/AOG.0000000000001405
38. ACOG. ACOG Clinical Practice Update: Paternal and Fetal Genotyping in the Management of Alloimmunization in Pregnancy. https://journals.lww.com/greenjournal/abstract/2024/08000/acog_clinical_practice_update__paternal_and_fetal.34.aspx
39. ACOG. Rho(D) Immune Globulin Shortages. https://www.acog.org/clinical/clinical-guidance/practice-advisory/articles/2024/03/rhod-immune-globulin-shortages
40. ACOG. Committee Opinion No. 691: Carrier Screening for Genetic Conditions. Obstetrics and gynecology. Mar 2017;129(3):e41-e55. doi:10.1097/aog.0000000000001952
41. ISPD. Joint Position Statement from the International Society for Prenatal Diagnosis (ISPD), the Society for Maternal Fetal Medicine (SMFM), and the Perinatal Quality Foundation (PQF) on the use of genome-wide sequencing for fetal diagnosis. Prenat Diagn. 2018;38(1):6-9. doi:10.1002/pd.5195
42. Palomaki GE, Chiu RWK, Pertile MD, et al. International Society for Prenatal Diagnosis Position Statement: cell free (cf)DNA screening for Down syndrome in multiple pregnancies. Prenat Diagn. 2021;41(10):1222-1232. doi:10.1002/pd.5832
43. Sandler SG, Flegel WA, Westhoff CM, et al. It's time to phase in RHD genotyping for patients with a serologic weak D phenotype. College of American Pathologists Transfusion Medicine Resource Committee Work Group. Transfusion. 2015;55(3):680-9. doi:10.1111/trf.12941
44. Regan F, Veale K, Robinson F, et al. Guideline for the investigation and management of red cell antibodies in pregnancy: A British Society for Haematology guideline. Transfus Med. Feb 2025;35(1):3-23. doi:10.1111/tme.13098
Coding
| CPT |
Code Description |
| 81171 |
AFF2 (ALF transcription elongation factor 2 [FMR2]) (eg, fragile X intellectual disability 2 [FRAXE]) gene analysis; evaluation to detect abnormal (eg, expanded) alleles |
| 81172 |
AFF2 (ALF transcription elongation factor 2 [FMR2]) (eg, fragile X intellectual disability 2 [FRAXE]) gene analysis; characterization of alleles (eg, expanded size and methylation status) |
| 81200 |
ASPA (aspartoacylase) (eg, Canavan disease) gene analysis, common variants (eg, E285A, Y231X) |
| 81209 |
BLM (Bloom syndrome, RecQ helicase-like) (eg, Bloom syndrome) gene analysis, 2281del6ins7 variant |
| 81241 |
F5 (coagulation factor V) (eg, hereditary hypercoagulability) gene analysis, Leiden variant |
| 81242 |
FANCC (Fanconi anemia, complementation group C) (eg, Fanconi anemia, type C) gene analysis, common variant (eg, IVS4+4A>T) |
| 81243 |
FMR1 (fragile X messenger ribonucleoprotein 1) (eg, fragile X syndrome, X-linked intellectual disability [XLID]) gene analysis; evaluation to detect abnormal (eg, expanded) alleles |
| 81244 |
FMR1 (fragile X messenger ribonucleoprotein 1) (eg, fragile X syndrome, X-linked intellectual disability [XLID]) gene analysis; characterization of alleles (eg, expanded size and promoter methylation status) |
| 81251 |
GBA (glucosidase, beta, acid) (eg, Gaucher disease) gene analysis, common variants (eg, N370S, 84GG, L444P, IVS2+1G>A) |
| 81252 |
GJB2 (gap junction protein, beta 2, 26kDa, connexin 26) (eg, nonsyndromic hearing loss) gene analysis; full gene sequence |
| 81253 |
GJB2 (gap junction protein, beta 2, 26kDa, connexin 26) (eg, nonsyndromic hearing loss) gene analysis; known familial variants |
| 81255 |
HEXA (hexosaminidase A [alpha polypeptide]) (eg, Tay-Sachs disease) gene analysis, common variants (eg, 1278insTATC, 1421+1G>C, G269S) |
| 81257 |
HBA1/HBA2 (alpha globin 1 and alpha globin 2) (eg, alpha thalassemia, Hb Bart hydrops fetalis syndrome, HbH disease), gene analysis; common deletions or variant (eg, Southeast Asian, Thai, Filipino, Mediterranean, alpha3.7, alpha4.2, alpha20.5, Constant Spring) |
| 81260 |
IKBKAP (inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein) (eg, familial dysautonomia) gene analysis, common variants (eg, 2507+6T>C, R696P) |
| 81290 |
MCOLN1 (mucolipin 1) (eg, Mucolipidosis, type IV) gene analysis, common variants (eg, IVS3-2A>G, del6.4kb) |
| 81329 |
SMN1 (survival of motor neuron 1, telomeric) (eg, spinal muscular atrophy) gene analysis; dosage/deletion analysis (eg, carrier testing), includes SMN2 (survival of motor neuron 2, centromeric) analysis, if performed |
| 81330 |
SMPD1(sphingomyelin phosphodiesterase 1, acid lysosomal) (eg, Niemann-Pick disease, Type A) gene analysis, common variants (eg, R496L, L302P, fsP330) |
| 81400 |
Molecular pathology procedure, Level 1 (eg, identification of single germline variant [eg, SNP] by techniques such as restriction enzyme digestion or melt curve analysis) |
| 81401 |
Molecular pathology procedure, Level 2 (eg, 2-10 SNPs, 1 methylated variant, or 1 somatic variant [typically using nonsequencing target variant analysis], or detection of a dynamic mutation disorder/triplet repeat) |
| 81403 |
Molecular pathology procedure, Level 4 (eg, analysis of single exon by DNA sequence analysis, analysis of >10 amplicons using multiplex PCR in 2 or more independent reactions, mutation scanning or duplication/deletion variants of 2-5 exons) |
| 81404 |
Molecular pathology procedure, Level 5 (eg, analysis of 2-5 exons by DNA sequence analysis, mutation scanning or duplication/deletion variants of 6-10 exons, or characterization of a dynamic mutation disorder/triplet repeat by Southern blot analysis) |
| 81405 |
Molecular pathology procedure, Level 6 (eg, analysis of 6-10 exons by DNA sequence analysis, mutation scanning or duplication/deletion variants of 11-25 exons, regionally targeted cytogenomic array analysis) |
| 81406 |
Molecular pathology procedure, Level 7 (eg, analysis of 11-25 exons by DNA sequence analysis, mutation scanning or duplication/deletion variants of 26-50 exons)) |
| 81412 |
Ashkenazi Jewish associated disorders (eg, Bloom syndrome, Canavan disease, cystic fibrosis, familial dysautonomia, Fanconi anemia group C, Gaucher disease, Tay-Sachs disease), genomic sequence analysis panel, must include sequencing of at least 9 genes, including ASPA, BLM, CFTR, FANCC, GBA, HEXA, IKBKAP, MCOLN1, and SMPD1 |
| 81443 |
Genetic testing for severe inherited conditions (eg, cystic fibrosis, Ashkenazi Jewish-associated disorders [eg, Bloom syndrome, Canavan disease, Fanconi anemia type C, mucolipidosis type VI, Gaucher disease, Tay-Sachs disease], beta hemoglobinopathies, phenylketonuria, galactosemia), genomic sequence analysis panel, must include sequencing of at least 15 genes (eg, ACADM, ARSA, ASPA, ATP7B, BCKDHA, BCKDHB, BLM, CFTR, DHCR7, FANCC, G6PC, GAA, GALT, GBA, GBE1, HBB, HEXA, IKBKAP, MCOLN1, PAH) |
| 81479 |
Unlisted molecular pathology procedure |
| 81599 |
Unlisted multianalyte assay with algorithmic analysis |
| S3844 |
DNA analysis of the connexin 26 gene (GJB2) for susceptibility to congenital, profound deafness |
| S3845 |
Genetic testing for alpha-thalassemia |
| S3846 |
Genetic testing for hemoglobin E beta-thalassemia |
| S3849 |
Genetic testing for niemann-pick disease |
| 0400U (effective 07/01/2023) |
Obstetrics (expanded carrier screening), 145 genes by next-generation sequencing, fragment analysis and multiplex ligation-dependent probe amplification, DNA, reported as carrier positive or negative |
| 0449U (effective 04/01/2024) |
Carrier screening for severe inherited conditions (eg, cystic fibrosis, spinal muscular atrophy, beta hemoglobinopathies [including sickle cell disease], alpha thalassemia), regardless of race or self-identified ancestry, genomic sequence analysis panel, must include analysis of 5 genes (CFTR, SMN1, HBB, HBA1, HBA2) Proprietary test: UNITY Carrier Screen™ Lab/Manufacturer: BillionToOne Laboratory, BillionToOne, Inc |
| 0488U (effective 10/01/2024) |
Obstetrics (fetal antigen noninvasive prenatal test), cellfree DNA sequence analysis for detection of fetal presence or absence of 1 or more of the Rh, C, c, D, E, Duffy (Fya), or Kell (K) antigen in alloimmunized pregnancies, reported as selected antigen(s) detected or not detected Proprietary test: UNITY Fetal Antigen™ NIPT Lab/Manufacturer: BillionToOne Laboratory, BillionToOne, Inc |
| 0489U (effective 10/01/2024) |
Obstetrics (single-gene noninvasive prenatal test), cellfree DNA sequence analysis of 1 or more targets (eg, CFTR, SMN1, HBB, HBA1, HBA2) to identify paternally inherited pathogenic variants, and relative mutation-dosage analysis based on molecular counts to determine fetal inheritance of maternal mutation, algorithm reported as a fetal risk score for the condition (eg, cystic fibrosis, spinal muscular atrophy, beta hemoglobinopathies [including sickle cell disease], alpha thalassemia) Proprietary test: UNITY Fetal Risk Screen™ Lab/Manufacturer: BillionToOne Laboratory, BillionToOne, Inc |
| 0494U (effective 10/01/2024) |
Red blood cell antigen (fetal RhD gene analysis), next-generation sequencing of circulating cell-free DNA (cfDNA) of blood in pregnant individuals known to be RhD-negative, reported as positive or negative Proprietary test: Rh Test Lab/Manufacturer: Natera™ |
| 0536U (effective 04/01/2024) |
Red blood cell antigen (fetal RhD), PCR analysis of exon 4 of RHD gene and housekeeping control gene GAPDH from whole blood in pregnant individuals at 10+ weeks gestation known to be RhD negative, reported as fetal RhD status Proprietary test: Prenatal Detect RhD Lab/Manufacturer: Devyser Genomic Laboratories, Devyser AB |
| S3845 | Genetic testing for alpha-thalassemia |
| S3846 | Genetic testing for hemoglobin E beta-thalassemia |
| S3849 | Genetic testing for niemann-pick disease |
Procedure and diagnosis codes on Medical Policy documents are included only as a general reference tool for each policy. They may not be all-inclusive.
This medical policy was developed through consideration of peer-reviewed medical literature generally recognized by the relevant medical community, U.S. FDA approval status, nationally accepted standards of medical practice and accepted standards of medical practice in this community, and other nonaffiliated technology evaluation centers, reference to federal regulations, other plan medical policies and accredited national guidelines.
"Current Procedural Terminology © American Medical Association. All Rights Reserved"
History From 2002 Forward
| 07/29/2025 | Annual review, no change to policy intent. Updating description, table of terminology, rationale and references. Adding CPT 81252, 81253, and S3844. |
| 02/26/2025 | Adding code 0536U effective 04/01/2025 |
| 09/05/2024 | Updated CPT coding. Added codes 0488U, 0489U and 0494U (effective 10/01/2024). No change in policy intent. |
| 08/19/2024 | Annual review, criteria #3 and criteria #5 reworded for clarity. Criteria #7 added stating: To screen single-gene mutations (i.e. autosomal recessive, autosomal dominant, X-lined) in the fetus, the use of non-invasive prenatal screening (NIPS) is not medically necessary. Updating description, note #2, rationale, references, table of terminology and updating coding verbiage. |
| 03/26/2024 | Updating policy section. Adding coding 0449U (effective 04/01/2024) No other changes made. |
| 07/31/2023 | Annual review, updating policy for clarity and consistency. Adding verbiage regarding Tier 1/2/3 screening. Also updating description, table of terminology, rational, and references, note and coding adding 0400U. |
| 06/14/2023 | Added code 0400U effective 07/01/2023. |
| 08/04/2022 | NEW POLICY |
.